Computational Characterization of MIPS in Camellia sinensis and it's Phylogenetic Implication
Anjan Hazra1,2, Nirjhar Dasgupta1, Chandan Sengupta2 and Sauren Das1*
1Agricultural and Ecological Research Unit, Indian Statistical Institute, India
2Department of Botany, University of Kalyani, India
Submission: June 28, 2017; Published: August 03, 2017
*Corresponding author: Sauren Das, Agricultural and Ecological Research Unit, Indian Statistical Institute, 203, Barrackpore Trunk Road, Kolkata 700108, India, Email: sauren@isical.ac.in
How to cite this article: Hazra A, Dasgupta N, Sengupta C, Das S. Computational Characterization of MIPS in Camellia sinensis and itís Phylogenetic Implication. Int J cell Sci & mol biol. 2017; 2(5): 555597. 10.19080/IJCSMB.2017.02.555597.
Abstract
Myo-inositol-1-phosphate synthase (MIPS) is a fundamental enzyme in sugar metabolism pathway, related to inositol in all living system. Inositol and its derivative compounds are involved in different signaling pathways and stress tolerance. Plant MIPS are dynamic in signal transduction, growth regulation, osmo regulation, abiotic and biotic stress response etc. Present study targets an in silico characterization of the MIPS from tea and estimate its phylogenetic status. The physicochemical characters and predicted quaternary structure of the protein had been analyzed using different bioinformatics tools. Predictive sub cellular localization is in line with the earlier works. All sequences were 80-90% identical among themselves, however MIPS3 isoform from Arabidopsis thaliana had larger similarity with tea MIPS than those of MIPS1 and 2. In a number of sites tea MIPS and ATMIPS3 had similar residue and different from other two, whereas in few sites tea had unique residue composition as well. More detailed analysis would help to understand the impact of single residue changes at specific site of differential function of the protein and its role in basic or stress compatibility of tea plant.
Keywords: Camellia sinensis; MIPS; Phylogenetic analysis; Physicochemical properties; Quaternary structure
Introduction
Myo-inositol-1-phosphate synthase (MIPS) is key enzyme in the synthesis of inositol related sugar metabolism in all living system. Inositol and its derived compounds are involved in different signaling pathways like, auxin transport, stress tolerance [1], oligosaccharide synthesis [2-4], cell death regulation, cell wall biosynthesis [5-9]. In plants, they play active role in signal transduction, growth regulation, osmoregulation, abiotic and biotic stress response etc [10-12]. Especially, they possess salt tolerance ability through protection of cellular structures from reactive oxygen species and control cellular turgor pressure [13,14]. However crystallographic structure of MIPS from higher plants, particularly in angiosperms, is still yet to be conclude about the structre-function relationship and its variation among different members. Recently in silico methods to determine structural and functional characteristics of a gene, transcript or protein is being accelerated to avoid of the wet lab experimental limitations and it works too [15-19]. Tea plant, Camellia sinensis (L.) O. Kuntze, is the potential source of health beverage and most popular non-alcoholic drink across the world and being an evergreen shrub it grows well in diverse habitat and wide range of environment [20].The potential of its existing stress combating ability in some verities is the point of interest of the scientist to understand its mechanism and in progress in future cultivars. Present study aims to characterize in silico an important stress related protein, MIPS from tea and its phylogenetic status.
Methods
Complete amino acid sequence of Camellia sinensis MIPS (Accession No. AJO70149.1) was retrieved through BLASTp program [21] at NCBI using a previously reported homolog from Porteratia coarctata [10] as query sequence. Camellia sinensis MIPS was computationally predicted via homology modelling with the python based modeler 9.12 tool [22] after selecting the most suitable template at protein data bank (Template id- 1LA2, Source - Saccharomyces cerevisiae). Essential ligand NAD+ was allowed to bind at each chain of the homotetrameric protein. The resultant pdb model was analysed for it's in silico quality checking and physicochemical property estimation. Using different bioinformatics tools Ramachandran plot [23], Z-score [24], subcellular localization [25], half-life, instability and aliphatic index [26] were estimated.
For a phylogenetic insight corresponding MIPS sequence was compared with the all annotated MIPS variety of Arabidopsis thaliana available at TAIR10 [27]. Multiple sequence alignment file was observed with the Boxshade server and represented accordingly. Multiple sequence alignment among the selected varieties was manually examined and a neighbor joining tree was drawn with the help of MEGA 7.0 software [28] to know about its isoform similarity. Relative divergent time was also calculated within the same tool.
Result and Discussion
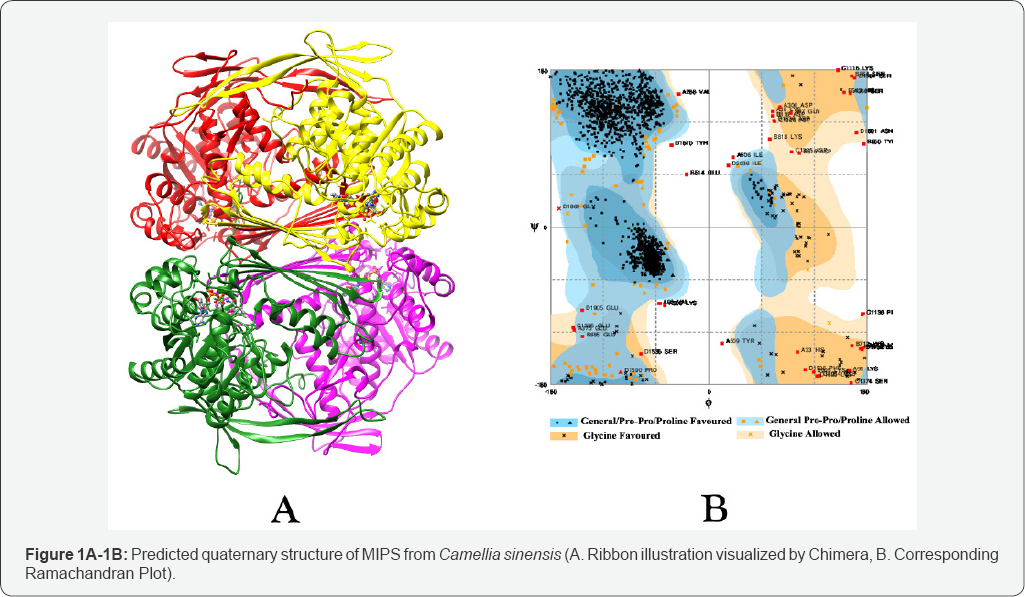
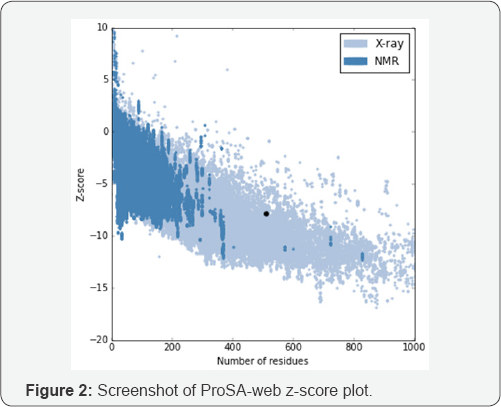
The BLAST returned homolog of MIPS isoform in tea was composed of 510 amino acid residues as found in others. Such polypeptide chain has an estimated half-life of 30 hours (mammalian reticulocytes, in vitro), >20 hours (yeast, in vivo) and >10 hours (Escherichia coli, in vivo) with the extinction coefficient is 52955M-1cm-1, at 280nm measured in water, Absorbance 0.1% (=1g/l)0.942, assuming all pairs of Cys residues form cystines and extinction coefficient is 52830M-1cm-1, at 280nm measured in water, Abs 0.1% (=1g/l) 0.940, assuming all Cys residues are reduced. The instability index is computed to be 31.59 which indicates the protein is stable. Moreover, the aliphatic index 90.92 and Grand average of hydropathicity (GRAVY) was calculated as -0.163. Quaternary structure of the protein was predicted using ligand modelling in Modeller 9.12 and visualized in Chimera [29] (Figure 1A). Ramachandran Plot analysis (Figure 1B) shown 93.4% of total number of residues in favored region and 4.4% in allowed region. Same trend of scores was found in earlier study conducted with modelled MIPS protein among different group of plants [30]. The z-score indicates overall model quality is displayed in a plot that contains the z-scores of all experimentally determined protein structures from different sources (X-ray, NMR) [24]. In predicted tea MIPS the value is calculated as -7.9 which also within the range of scores typically found for native proteins of similar size (Figure 2). According to CELLO prediction the protein was predicted to be localized in Periplasmic and Cytoplasmic regions which is similar with earlier report of its localization [4,14].

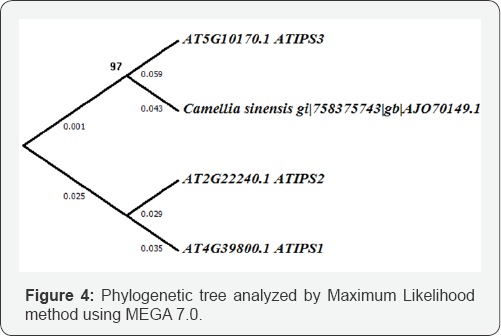
Multiple sequence alignment of Arabidopsis thaliana MIPS varieties with the model protein in present study (Figure 3) revealed some important findings. All sequences were 80-90% identical among themselves, however MIPS3 isoform had larger similarity with tea MIPS than other two. In a number of sites tea MIPS and ATMIPS3 had similar residue which largely differ from other two, whereas in few sites tea had unique residue composition too. It is also reflected in the phylogenetic tree (Figure 4) where tea MIPS and ATMIPS3 belonged to same clade and has common ancestor to the divergent MIPS1 and MIPS2 isoforms. According to relative divergence time scale MIPS1 and MIPS2 clade took higher time to evolve than that of MIPS3 clade and tea MIPS is much primitive than MIPS3 itself. More detailed analysis would be of help to understand the impact of single residue changes at specific site of differential function of the protein and its role in basic or stress induced state of the tea plant.
Acknowledgement
The author, A. Hazra is thankful to National Tea Research Foundation, Tea Board, India, for providing research fellowship.
References
- Taji T, Takahashi S, Shinozaki K (2006) Inositols and their metabolites in abiotic and biotic stress responses, in Biology of Inositols and Phosphoinositides. Springer 39: 239-264.
- Karner U (2004) Myo-Inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J Exp Bot 55(405): 1981-1987.
- Michell RH (2007) Evolution of the diverse biological roles of inositols. Biochem Soc Symp 74: 223-246.
- Donahue JL (2010) The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22(3): 888-903.
- Loewus FA, Murthy PP (2000) Myo-Inositol metabolism in plants. Plant science 150(1): 1-19.
- Stevenson JM, Perera IY, Heilmann I, Persson S, Boss WF (2000) Inositol signaling and plant growth. Trends Plant Sci 5(6): 252-258.
- Downes CP, Gray A, Lucocq JM (2005) Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol 15(5): 259-268.
- Abreu EF, Aragao FJ (2006) Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulisf. flavicarpa) expressed during seed development and environmental stress. Ann Bot 99(2): 285-292.
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446(7136): 640-645.
- Ray S, Patra B, Das-Chatterjee A, Ganguli A, Majumder AL (2010) Identification and organization of chloroplastic and cytosolic L-myo- inositol 1-phosphate synthase coding gene (s) in Oryza sativa: comparison with the wild halophytic rice, Porteresia coarctata. Planta 231(5): 1211-1227.
- Zhai H, Wang F, Si Z, Huo J, Xing L, et al. (2016) A myo-inositol-1- phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol J 14(2): 592-602.
- Kaur H, Shukla RK, Yadav G, Chattopadhyay D, Majee M (2008) Two divergent genes encoding L-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ 31(11): 1701-1716.
- Nelson DE, Rammesmayer G, Bohnert HJ (1998) Regulation of cell- specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 10(5): 753-764.
- Khurana NH, Chauhan, Khurana P (2012) Expression analysis of a heat-inducible, Myo-inositol-1-phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant cell reports 31(1): 237-251.
- Hazra A (2015) Computational fishing and structural analysis of MIPS protein from two important plant groups. International Letters of Natural Sciences 42.
- Cichero E, Stefano E, Michele T, Silvia F, Andrey SG, et al. (2016) A homology modelling-driven study leading to the discovery of the first mouse trace amine-associated receptor 5 (TAAR5) antagonists. Med Chem Comm 7(2): 353-364.
- Smith AA, Caruso A (2013) In silico characterization and homology modeling of a cyanobacterial phosphoenolpyruvate carboxykinase enzyme. Structural Biology 2013.
- Nagarajan SK, Madhavan T (2016) Protein Phosphatase 1D (PPM1D) Structure Prediction Using Homology Modeling. J Chosun Natural Sci 9(1): 35-40.
- Dasgupta N, Hazra A, Bhattacharya S, Das S (2017) In Silico screening of Putative miRNAs and their Targets from a Common Mangrove Bruguiera gymnorrhiza. International Journal of Cell Science & Molecular Biology 2(1): 1-18.
- Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, et al., (2017) The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol Plant 10(6): 866-877.
- Altschul SF (1990) Basic local alignment search tool. Journal of molecular biology 215(3): 403-410.
- Webb B, Sali A (2014) Protein structure modeling with MODELLER. Methods Mol Biol 1137: 1-15.
- Lovell SC, Davis IW, Arendall WB, Richardson DC, Word JM, et al. (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50(3): 437-450.
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(suppl_2): W407-W410.
- Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64(3): 643-651.
- Gasteiger E (2005) Protein identification and analysis tools on the ExPASy server. Springer.
- Swarbreck D (2007) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36(suppl 1): D1009-D1014.
- Kumar S, Stecher G, Tamura K, (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7): 1870-1874.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25(13): 1605-1612.
- Hazra A, Nandy P (2016) Myo-inositol 1-phosphate synthase-the chosen path of evolution. BioTechnologia. Journal of Biotechnology Computational Biology and Bionanotechnology 97(2): 95-108.






























