Molecular Imaging with Chemokine Receptor4 (Cxcr4) In Breast Cancer
Yadav Ajay Kumar, Pandit Anil and Ansari Quamrul Haque*
Department of Radio-diagnosis Imaging and Nuclear Medicine, BP Koirala Memorial Cancer Hospital, Nepal
Submission: December 17, 2016; Published: January 17, 2017
*Correspondence Address: Ansari Quamrul Haque, Department of Radio-diagnosis Imaging and Nuclear Medicine, B.P. Koirala Memorial Cancer Hospital, Bharatpur, Chitwan, Nepal, Email: kamrulhak@gmail.com
How to cite this article: adav A ,Pandit A, Ansari Q. Molecular Imaging with Chemokine Receptor4 (Cxcr4) In Breast Cancer. IInt J cell Sci & mol biol. 2017; 1(4) : 555568. DOI:10.19080/IJCSMB.2017.01.555568
Abstract
In this study, CXCR4 siRNA labeled with 99mTc was used in gene imaging of nude mice for the breast carcinoma. Small interference RNA (siRNA) targeting human CXCR4 was radiolabeled by using the bifunctional chelator HYNIC. Negative control siRNA were synthesized by siRNA targeted to CXCR4 mRNA and radiolabeled by 99mTc with the bifunctional chelator HYNIC. Animal models of nude mice bearing human breast cancer MDA-MB-231 were established, divided them into three groups, 10 mice in each group. 7.4MBq of 99mTcO4-, 99mTc- HYNIC(siRNA) and 99mTc-HYNIC(-siRNA) were separately injected through tail vein to the three groups. Images of these animal modelswere acquired with SPECT, the ratio of T/M was calculated by region of interest (ROI). The data were analyzed by two-sample t test and analysis of variance. Imagesof tumor sites at 1hr have different degree of radionuclide accumulation, which interference group is most obvious, and as the extension of time, the accumulation of interference probe in tumor tissue was gradually increased. At 1, 4 and 10hr after injection, there were statistically significant differences in different groups.
The T/M ratios in 99mTcO4-group and 99mTc-HYNIC(-siRNA)-group were significantly lower than 99mTc -HYNIC(siRNA)-group at 1, 4, 10hr respectively (tcontrol = 29.20, 33.84 and 38.07, t99mTcO4- = 31.29, 37.61, 41.86, all P <0.01), and T/M ratios at 1h, 4h , 10h of 99mTc -HYNIC(- siRNA)-group and 99mTcO4-groups were not significantly different (t = 0.392, 0.318 and 0.533, all P> 0.05). The imaging with 99mTc- HYNIC- siRNA may be a promising method for diagnosis of breast cancer.
Keywords: Chemokine receptor-4 (Cxcr4); Small interference RNA (Sirna); Negative groups of small interference RNA (-Sirna); Mda-Mb-231 Cells; 99mtc
Introduction
Nuclear Medicine is an important branch of a molecular imaging, occupies an important position, and has broad prospects for development. Nuclear Medicine is the imaging technique in which use of radiopharmaceutical in-vivo, revealing the physiological, biochemical and metabolic changes at the molecular level of the human body. It helps us to know physiological and pathological processes living body at the molecular level. It is Non-invasive, real-time imaging; quantitative and qualitative research of the biological behavior provides information on the molecular level and helps in treatment of disease. It reveals abnormal changes in living body, occurrence and development of disease and evaluation of drug efficacy in diseases. Development of nuclear medicine, it plays important role in the early diagnosis and treatment of neoplastic diseases, along with rapid development of PET, PET/CT, PET/ MRI, SPECT/CT, SPECT/MRI, nuclear medicine has become another highlights.
Gene probes labeled imaging is a vivo imaging method which is used in the gene or protein level of tumor-specific gene expression and will be a non-invasive detection of tumor, including antisense imaging, exogenous gene introduced imaging and multidrug resistance (MDR) gene imaging. Antisense imaging is based on the principle of complementary base pairing in which open radionuclide-labeled synthetic antisense probe is use target mRNA or target gene. Gene sequences cancer cells antisense probe with specific binding is done with vitro imaging instrument which display gene or gene over expression organizational, thereby forming a new diagnostic method. Exogenous gene transduction imaging is based imaging suicide therapy in which the target genes or target substrates conducted. Introducing the expression levels in tumor cells, sensitive genes determine tumor sensitivity to the drug so transduced gene imaging has significant value in guiding cancer gene therapy and evaluate the efficacy of gene therapy.
MDR tumor cells at the same time are a function of a number of structurally different cytotoxic compounds simultaneously exhibits the phenomenon of tolerance. Tumor cell MDR phenomenon is due to the over-expression of the gene encoding MDR-phosphate glycoproteins and MDR gene related proteins due. MDR gene phosphate glycoproteins and MDR-associated protein gene can be diagnosed by 99mTc-MIBI and some cationic lipophilic compounds cleared extracellular. 99mTc-MIBI retention in tumor cells and the amount of phosphoric acid glycoprotein in MDR genes and gene-related protein is expressed. The expression was negatively correlated with 99mTc-MIBI and used to evaluate certain tumor MDR cases. In this paper, some basic problems of siRNA imaging technology and the current status of tumor antisense imaging is briefly overviewed.
RNA interference (RNAi) is a double-stranded small interfering RNA (siRNA) to make specific mRNA degradation, thereby enabling the gene transcriptional silencing of a phenomenon [1]. Fire et al. [2] found that the injection of the same dose of double-stranded RNA as compared with singlestranded RNA, efficiency of double-stranded is 10-100 times than single-stranded RNA. The aim of their study to suppress gene expression behavior known as RNA interference (RNAi), trigger RNA interference phenomenon called small interfering RNA. So in 2006 they won the Nobel Prize in Physiology/Medicine. Since then, a new gene technology i.e. RNA interference technology has emerged. Using their specific nucleotide sequence, according to the base pairing rules, the antisense strand or sense strand mRNA specific for binding to the complementary region, formation of a conjugate with the RISC, Argonaute2 degradation purposes mRNA, the degradation of the remaining fragments of RNA hydrolysis by the intracellular hydrolytic enzymes [3], in order to achieve the purpose of regulation of gene expression.
Scintigraphy with radionuclides labeled with siRNA is based on the principle of complementary base pairing, so that the target gene with small interfering RNA specific binding, gene expression can dynamically monitor the overall level of the disease tissue. In theory, as long as the target organ of memory in some kind of DNA or mRNA over expression, small interfering RNA molecules can be synthesized specific radiolabeled probes, at the molecular level in order to monitor overall changes in gene expression. Therefore, the main problems of small interfering RNA imaging technology applied is determined to play a role in tumor genesis dominant oncogene. Although there have been discovered more than 100 cancer genes, but only a portion of oncogenes involved in tumor process occurs [4].
Choose a reasonable interference sequence is a prerequisite for the successful application of RNA interference technology, but also one of the most basic, the most critical step. The siRNA must pass the gene sequence and the gene of interest to make binding and degradation by Argonaute-2, therefore, the key lies in siRNA sequences highly homologous genes and other non- homologous gene. The sequence is to determine the choice of specific RNA interference of the key, but also the basic principles of siRNA design. How to filter out the siRNA sequences from different silencing efficiency in the most efficient sequence of interference requires rigorous design and constantly test [5]. First, from the downstream gene initiation codon 50100 nucleotides, has been to the stop codon between 50-100 nucleotides upstream area looking for the ideal siRNA sequence, the closer to the target gene 3'end Gene silencing effect may be better [6]. Our preliminary experimental results are consistent with this hypothesis. Second, there are reports that the general should be two siRNA sequences adenine (AA) as a starting sequence to end UU base for the final, has been shown that the sequence of siRNA to AA UU starting when the end of the target gene has the highest inhibition efficiency [7]. But there are also some non-AA literature says smaller off-target effects of siRNA starting, so now is not recommended as starting with AA.
Third, since the promoter transcription termination sequence of 4 to 6 consecutive T, is continuously greater than or equal to 4 or T 4 A sequence of the gene should be avoided in the object, the continuous or greater 4 T or sequence of four A's, if present in the sequence inside, it may cause premature termination of transcription. It should avoid more than three consecutive G, because polyG sequence can overlap to form a block structure, seriously affecting the interference effect of siRNA. Fourth, siRNA sequence length should be 21-23 bases [8], there are 27 or 19 bases recommended that the inhibitory activity greater than 21 bases, induce interferon little reaction, from the low efficiency of concentration and so on. However, in the present study, 21 bases have been a good result. Fifth, the G siRNA sequences C content is very important, lower G/C ratio of siRNA sequence of the gene silencing effect better [9], G, C base amount of 30% to 70% can be used as an alternative. Finally, the continuous and single base inverted repeats should try to avoid in the siRNA sequence [10], because the body will make a continuous sequence of siRNA stability is reduced, thereby reducing the interference of siRNA. The siRNA sequence palindrome or repeat sequences may form a hairpin structure, so the declining role of siRNA in effective concentration.
It can further select the appropriate genetic sequences [11]. The purpose is according to the following criteria, and to improve the success rate of the target gene siRNA design. Criteria;
- G/C content of about 30% ~ 52%;
- Sense strand 3, the end of 15 to 19 bases in A/T (U) is appropriate, should contain 3 or 3 or more A/T (U);
- Repeat or palindromic sequence should be present in the chain, otherwise the sense strand or antisense strand itself may form a hairpin structure, the impact of siRNA duplexes formed. T value chain within melting temperature the sequence can be used to estimate fat Trend folder structure formation, should try to choose a sequence T is less than 20%;
- 19 Preferred A sense strand;
- 3 Preferred A sense strand;
- 19 sense strand;
- 10 preferred U justice chain not a G or C;
- 13 sense strand cannot be G. 1,2,3 reflects standard temperature dynamic characteristics of siRNA, may be associated with small interfering RNA and RISC recognition and binding, the standard 4,5,6 representing siRNA nucleotide sequence-specific, they may be small interfere with the interaction between RNA and protein related. It was calculated based on the eight criteria: 1,3,4,5,6 to meet the standards when each plus 1 point, each minus 1 point when 7-8 is not satisfied, for the second criterion, the sense strand of the 3, end 15 When ~ 19 bases each containing an A or U will add 1 point. Score more than 6 points; compared with siRNA may have functions.
In imaging studies, in vivo stability of siRNA is a very critical issue, but also a basic requirement, because if siRNA has a very good stability, it conducts effective imaging studies. Extracellular, siRNA degradation very quickly, by intravenous injection of free siRNA can easily be RNase enzyme recognition further degradation [3], half-life in the plasma less than 30 minutes, is not part of the degradation are scattered throughout the body. In order to increase the stability of siRNA in vivo, people were modified chemically to maximize the reduction of its sensitivity to RNase, conducted a variety of chemical modifications including RNA backbone modifications (such as phosphorothioate and borane modified phosphoric acid), 2 'modified ribose, 3' or 5 'end modification and the like.
Experiments confirmed that siRNA chemically modified for resistance to nuclease significantly enhanced, and some siRNA modified after the interference of the mRNA activity also strengthened.
In recent years, some chemical modification methods have been successfully used to improve the stability of RNA molecules, [12] including phosphorothioate (phosphorothioate, PS) and 2'-methoxy modifications [13]. 2'-OMe RNA modification can not only improve the resistance to a variety of physical and chemical factors, while a variety of enzymes to resist degradation capability has also been enhanced, but also improve the transport capacity in vivo RNA molecules. Therefore, this experiment uses a chemical modification of 2'-OMe, select one target chemokine receptor 4 mRNA in siRNA labeled as a probe target, siRNA construct 99mTc-labeled probe by coupling a bifunctional chelator HYNIC, to obtain a more ideal labeling efficiency and stability. On this basis, the researchers labeled probe siRNA in vivo biodistribution and imaging, and discuss its prospects in vivo imaging studies.
In gene imaging process, there is a problem that must be considered is the use of what radionuclides and what labeling method chosen. Among the many radionuclides, 111In has right half-life and energy peaks, very suitable for imaging, but their access needs accelerator, so expensive, the impact of its wider application. Radionuclides are currently widely used in clinical as 99mTc, physical half-life of about 6 hours, to peak 140keV, cheaper, convenient and simple to obtain, pure y-ray emission, suitable for imaging and many other advantages.
Currently 99mTc labeled with both direct and indirect labeling methods. Direct labeling i.e. between C atom labeled protein, polypeptide, or other substance is labeled with a radionuclide to form a covalent bond, but its more intense labeling conditions, are markers of damage, covalent bonding and weak easy to fall off and other shortcomings, it is now rarely used. Currently we pay more attention to the indirect labeling method, using a bifunctional chelating agent in which a radionuclide marker is connected with the mild reaction conditions, is a marker of small damage. Bifunctional chelating agent is used to avoid the many problems of direct labeling method, but also make the product of the nature of the basic requirements of radionuclide imaging. Bifunctional chelating agents plays an important role in the siRNA radioactive tags, have now produced for gene imaging probes. Bifunctional chelator have diethylenetriaminepentaacetic acid (DTPA), triethylene glycol mercaptoacetylglutamyl -N- hydroxysuccinimide ester (NHS-MAG3) and hydrazine Gini ancient amide (HYNIC), now more commonly used HYNIC and NHS-MAG3 two kinds.
Abrams MJ et al. [14], they synthesize HYNIC as an effective bifunctional chelating agent, started from the beginning of the application developed in foreign countries, and is widely used marker genes, proteins and antibodies like. So, HYNIC uses in research and development of experimental nuclear medicine imaging plays a very important role in promoting. In the course of radiolabeled using HYNIC often we need to add some synergistic ligand, more commonly used are N- tris(hydroxymethyl) methylglycine (tricine) and ethylenediamine diacetate (EDDA), because of HYNIC not occupy 99mTc the structure of all positions, other positions will need to be filled by co-ligands. Due to the structure of proteins and peptides vary, choose ligand marked rate impact [15], and therefore synergistic ligand specific markers should be different.
Radionuclide labeled chelator usually first coupled with the marker, then radiolabeled. When using HYNIC as a chelating agent, there is a higher rate of product marking; radiochemical purity after purification can reach more than 90%, and to obtain a higher specific activity. Studies have shown that when using 99mTc to be directly connected to the product labeling yields of 10% or less. Factors that affect the labeling yield analysis found that the main factors are the concentration of the labeled species, the molar ratio of coupling agent to be markers, collaborative ligand selection, marking time and temperature.
In 1997, Hnatowich DJ [16] team synthesized NHS-MAG3 [16], studies have confirmed that the use of NHS-MAG3 as a coupling agent for DNA, antisense oligonucleotides, peptide nucleic acids, small peptides and mixed backbone oligonucleotides with 99mTc, have a higher rate of mark and the mark rate is not affected by time and temperature. It is also confirmed that the radiochemical purity after purification can reach more than 90%, but do not use direct coupling agent was subjected to the above are labeled 99mTc labeled mark ratio of 5% or less. Serum stability studies show that the use of NHS-MAG3 as a coupling agent is marked outside the object has good stability.
Of course, the NHS-MAG3 and HYNIC have many similarities, such as NHS-MAG3 and HYNIC can be used for nucleic acids, proteins, antibodies, peptides and other coupling mark to give the product labeling efficiency, radiochemical purity and no significant difference. The main difference between the two at the following Table 1 [17].
The application of the two coupling agent with radionuclide 99mTc to nucleic acids, proteins, polypeptides, antibodies and other indirect labeled, to avoid damage, while labeling efficiency, radiochemical purity and specific activity were significantly increased . Currently HYNIC and NHS-MAG3 is widely used in nuclear medicine imaging studies play an important role in research for cancer, inflammation, cardiovascular system diseases, and widely used in the study of molecular imaging.
In the imaging studies, how to make the siRNA molecule labeled probe as much as possible after entering the target cells is another key issue. Imaging quality is closely related to the amount of the probe into the target cells, if the amount of the probe into the target cells less soft tissue corresponding background will increase, the lesions showed unclear, therefore, in order to improve transfer efficiency of the transporter probes people made a lot of research.
Liposome is also known as liposomes, is a novel formulation has targeted drug delivery capabilities. Vesicular structures for drug molecules are formed by the phospholipid bilayer membrane. Since the structure of biological membrane phospholipid bilayer is good biocompatibility. Compared to the use of liposome-mediated viral transduction has the following obvious advantages:
- The process of forming a complex with liposomes genes easier;
- Easy to mass production;
- Non-viral liposomal carrier with the cell membrane Fusion and the desired gene into the cells, followed by lipid degradation, non-immunogenic and non-toxic in a certain concentration range;
- Can protect the transducer so that it is not easy to nuclease degradation;
- Carried by liposomes, the purpose of the gene sequence may be transported to a target destination;
- Transfection process is simple and reproducible.
Also according to different needs, liposomes are prepared different in charge, size, flow, as well as pH. Now the new targeted liposomes are prepared, such as proliposomes, body length cycle liposomes, thermosensitive liposomes, immunoliposomes and pH-sensitive liposome. Liposomes can be extended siRNA molecule probes duration of action within the cell. Studies have shown that in liposome-mediated; siRNA molecule probe in the blood circulation is stable for a long time, compared to pure siRNA plasma half-life of less than 30min.
In 1970, it was found that Phagocytosis, receptor-mediated cell-specific transfer of exogenous substances into the intracellular one way. Receptor-mediated phagocytosis has two significant characteristics:
- high transfer efficiency [18,19];
Bcell, tissue or organ specificity. The basic steps are: sialic acid orosomucoid off on first use of an intermediate linker to asialoorosomucoid (ASOR) in combination with polylysine (PL) covalently resulting ASOR- PL nucleic acid complex is carried tools. At neutral pH environment, polylysine mixed with siRNA, forming ASOR-PL-AS-ODN complexes through a certain treatment, such complexes can be cell surface receptors that specifically recognize, then to be swallowed cells. This transfer approach has two significant advantages. First, receptors efficient and loyal, secondly, protective layer between the protection of the surrounding environment can be transferred nucleic acids, nuclease degradation of the environment has a certain ability to resist, can improve the transfer efficiency.
Breast cancer is a common malignancy in women, and its incidence in many countries and regions has been growing [20]. Numerous studies show that CXCR4 expression in at least 23 kinds of different types of cancer cells, such as breast cancer, ovarian cancer, prostate cancer, colorectal cancer and malignant melanoma [21,22]. Therefore, SiRNA can be labeled with radionuclide tracer to diagnose abnormal changes in early detection of breast cancer gene level and it will undoubtedly provide a reliable and effective method for the early diagnosis of breast cancer.
Materials and Methods
The main materials and equipment
CXCR4 -siRNA
Interference group CXCR4-37
'GGGACUAUGACUCCAUGAATT 3'
Antisense: 5’UUCAUGGAGUCAUAGUCCCTT3’
In the control group
Sense: 5 'UUCUCCGAACGUGUCACGUTT 3'
Antisense: 5'ACGUGACACGUUCGGAGAATT3'
In 5 antisense strand ‘end a six-carbon-hexyl primary amine and ammonia structure, as HYNIC coupled with a group of the 3’ end two used to improve the antisense nucleotide binding activity of the thymus deoxy d (TT ) structure.
Reagents
- MDA-MB-231 breast cancer cells
- HYNIC
- Lipofectamine 2000
- Stannous chloride (SnCl2)
- NH4HCO3
- N, N- dimethylformamide,
- Tricine
- Acetonitrile
- Sephadex G25
- C18 reverse phase column
- Na99mTcO4-
- SPECT: Duet Camera
- y-Counter,
- UV spectrophotometer
- NU/NU strains
Experimental Methods
The first step, the first Sep-Pak C18 reverse phase column pretreated. First with HPLC grade acetonitrile 10ml slow slowly pushed through the column, and then triple-distilled water 20ml was slowly pushed through the column. The second step, the pure HYNIC dissolved in DMF at a concentration of 10mg/ ml, taking siRNA 5OD (165µg), to 25mmol/L bicarbonate buffer (pH 8.5) was dissolved to a final concentration of 5µg/µl. The third step, in accordance with the siRNA: HYNIC molar ratio = 1:20, under the oscillation condition, the HYNIC solution was added drop wise freshly prepared siRNA bicarbonate buffer, RT for 60min, mixed with distilled water of siRNA with HYNIC was diluted to 1ml, and then the mixture through Sep-Pak C18 reverse phase column purification. The following step elution with eluent:
- 10ml 25mM ammonium bicarbonate solution elution time;
- 10ml containing 5% acetonitrile in 25mM ammonium bicarbonate solution eluted first.
- 10ml 5% aqueous acetonitrile twice.
- 1ml 30% acetonitrile pour in water four times. The effluent was collected, 8 drops per tube, 80, UV spectrophotometer absorbance of each tube in the combined peak tube at 260nm per tube 10µg aliquots (1OD = 33µg), - 20E storage backup. 10µg aliquot is stored siRNA-HYNIC, using 50µl concentration 25mmol/L of bicarbonate buffer solution. Then it is added to 100µl concentration 7mg/ml solution of Tricine, 10µl freshly prepared was dissolved in 0.1mol/L hydrochloric acid SnCl2 solution (1mg/ml) and 185MBq of 99mTcO4-. Mix the reaction at room temperature for 60min. After that using Sephadex G25 gel chromatography purification tag, mixture obtained after completion of normal saline as the eluent, to collect the liquid dripping, 6 drops per tube, a total of 80, measuring radioactive counts each tube and the absorbance at 260nm of each tube. CXCR4 siRNA interference group and control group siRNA in accordance with the above method and 99mTc labeled HYNIC coupling. Depicting 5OD (165µg) not coupled HYNIC interference group and control group CXCR4 siRNA, the same procedure as described above, respectively 99mTc labeled purified using Sephadex G25 chromatography analysis.
Radiochemical purity of labeled analyte
Labeled 10µg interference group and control group CXCR4 siRNA, One use strips of paper chromatography, the stationary phase of acetone and saline as measured double agent, application segmentation measurements obtained Tags rate and radiochemical purity of the product.
Stability analysis of markers
CXCR4 siRNA interference group and control group were labeled siRNA to 1.0µg/ml concentration was dissolved in PBS and fresh normal human serum in 37oC, respectively, incubated 30min and 120min, and then use the Sep-Pak C18 reversed phase chromatography column to analyze, measure the radioactivity of each collection tubes, each tube of the measured radioactivity distribution curve plotted radioactivity, radioactive peak was observed drift, to evaluate the stability of the markers.
Liposomes 2000 25µl and 475µl serum-free DMEM culture medium mix, allowed to stand for 5min, the 10µg 99mTc-HYNIC- siRNA was dissolved in 500µl of serum-free DMEM medium, and after the two-part liquid mix was allowed to stand 20 min. Group 99mTc-HYNIC-siRNA 10µg according to the same method liposome-encapsulated.
Cell culture and establish a nude mouse model of breast cancer
Open water bath temperature was adjusted to 37oC, in a sterile laboratory equipment needed for the stage gracefully, will be equipped with a liquid nitrogen tank from human breast cancer cells (MDA-MB-231) of the vials were rapidly removed and placed in 37oC warm water and continue to shake. After the cells were melted and 75% alcohol wipe disinfection and excesses in alcohol lamp above the flame, preferably with tweezers lid open frozen in sterile units in alcohol lamp burning above the flame lightly frozen pipe mouth, disposable pipette the cell suspension and transferred to sterile centrifuge tube by adding 2-3ml DMEM culture medium containing 10% calf serum (penicillin 100U/ml, streptomycin 100|ig/ml), mix by pipetting. 1200 r/min centrifugal 3min, the supernatant was discarded, adding an appropriate amount of 10% fetal bovine serum DMEM culture medium, the bottom of the tube with a straw pellet cells uniform, then transferred to a sterile disposable cell culture flasks, placed at 37oC, 5% CO2 incubator. 6-10 hours to observe the situation of adherent cells.
Breast cancer cells was changed
The flasks were removed from the incubator, placed under the microscope to observe the growth state of the cells, whether viable, such as the number of viable cells was observed, and the presence or absence of bacterial growth morphology. Sterile stage gracefully experimental equipment needed, discard the culture flasks of broth, adding an appropriate amount of saline gently washed two times discarded. Then add the right amount of DMEM containing 10% FBS culture medium, placed in an incubator to continue to foster growth in the number of cells to be the range of test requirements.
Breast cancer cells were passaged
Remove the cell culture flasks, placed in a state to observe the cells under a microscope. As well adherent cells grew well, the number of standard (density up to 90%), the cells were passaged. When Discard broth, adding an appropriate amount of saline gently washed two times, adding 0.25% trypsin 8 drops, digestive cells, placed under the microscope to be adherent cell morphology from spindle becomes round when, indicating cell it has been digested down from the sides of the bottle. Discard trypsin, adding 3-4ml DMEM culture medium containing 10% fetal calf serum, pipetting with a sidewall, the sidewall will blow from the next cell. Transferred to sterile tubes, 1200r/ min centrifugal 3min, the supernatant was discarded, adding an appropriate amount of DMEM containing 10% FBS culture medium, the bottom of the tube with a straw pellet cells Army uniform and evenly distributed in a sterile disposable cell culture flasks, each adding an appropriate amount broth uniform, placed conditions 37oC, 5% CO2 incubator.
Frozen breast cancer cells
Logarithmic growth phase cells, discard the broth, adding an appropriate amount of saline and gently rinse twice with 0.25% trypsin (same cells were passaged), discarded trypsin, containing 10% fetal bovine serum in DMEM 2 -4ml, hard wind and percussion with a straw adherent cells, the cells were down as much as possible from the sides of the bottle all the percussion. Transferred to sterile tubes, 1200r / min centrifugal 3min, the supernatant was discarded, adding 2-3ml cryopreservation fluid, straw gently pipetting the cell suspension evenly dispensed by adding sterile vials, the sealing tape, marking After placed in -80oC refrigerator for 24 hours, and finally placed in a liquid nitrogen tank storage.
Establishing nude mouse model of breast cancer
In the logarithmic phase of human breast cancer MDA- MB-231 cells were passaged cell method for producing single cell suspension cell count plate count, to calculate the total cell within the four corners of the large number of squares. Cells / ml = (total 4 large grid cells / 4) x 104. In serum-free DMEM culture medium the cell concentration was adjusted to 106-107 / ml for the experiments. On the outside of the right leg of nude mice inoculated subcutaneously with tumor cell suspension 0.2 ml, after inoculation of mice raised in a sterile environment, the growth of the tumor was observed every day until the tumor grew to a diameter at about 1 ~ 2cm, used in the experiment.
Imaging research
Randomly (simple random sampling) 30 breast tumor in nude mice were divided into three groups, each 10 mice were injected intravenously liposome free 99mTcO4-, interference 99mTc-HYNIC-siRNA group and control group 99mTc-HYNIC-siRNA (doses are 7.4 MBq). Mice mass fraction of 5% chloral hydrate anesthesia fixed, using low-energy collimator, to the peak 140 KeV, 25 x 256 matrix, 20% of the window width, after injection static imaging were taken 1h, 4h and 10h. Radioactivity counts were taken on ROI at each time and the contralateral, and the ratio (T/M) ratio were calculated.
Statistical analysis
Using Graph Pad Prism 5 software for data analysis, measurement data with x (_) ± s representation, P <0.05 when the difference was statistically significant.
Results
Characteristic mark rate of 99mTc-labeled products
Little by Sephadex G25 column gel analysis, interference HYNIC-siRNA group and control group HYNIC-siRNA 99mTc labeled after all there are two UV absorption peaks, of which the first peak is relatively high and wide, their radioactivity was measured and found only the first peak containing radioactive. Before and after the merger of the first peak several collection tubes, calculate the mark rate. HYNIC not coupled siRNA interference group and the control group of 99mTc labeled siRNA rate (4.91 ± 2.58)% and (4.82 ± 3.09)%, respectively, after the coupling HYNIC siRNA interference group and control group rate of 99mTc labeled siRNA were ( 61.26 ± 2.47)% and (60.85 ± 2.76)%, results showed HYNIC can significantly improve the rate of radiolabeled small interfering RNA. Interference 99mTc- HYNIC-siRNA group and control group 99mTc -HYNIC-siRNA radiochemical purity are more than 90%. By Sep- Pak C18 column analysis interference 99mTc-HYNIC-siRNA group and control group 99mTc -HYNIC-siRNA were dissolved in 37oC fresh human serum in PBS and the drift after 30min and 120min, no significant radioactivity peak, indicating mark product stability, good activity of the molecule.
Establish nude mouse model of breast cancer
After the logarithmic growth phase of human breast cancer MDA-MB-231 cells into a single cell suspension, the cell concentration was adjusted to 106-107/ml for experiments. On the outside of the right lower extremity female nude mice inoculated subcutaneously with tumor cell suspension 0.2ml, sterile environment, feeding about 3-4 weeks, tumor growth, tumor diameter length to be about 1-2cm, used in the experiment. As shown in Figure 1.
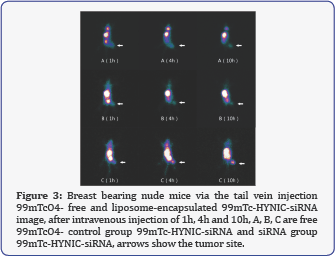
After intravenous injection of liposome free 99mTcO4-, interference 99mTc-HYNIC-siRNA group and control group 99mTc- HYNIC-siRNA (doses are 7.4 MBq) of nude mice bearing breast cancer, the mass fraction of 5% chloral hydrate anesthesia, at 1h, 4h and 10 h after injection of the imaging agent, respectively. Tumor and contralateral muscle techniques to measure ROI radioactivity count at each time point and calculate the ratio. Interference injection 99mTc-HYNIC-siRNA group of mice, after 1h the visible tumor radioactive gathered, 4h and 10h tumor imaging clearer, T/M ratio increased, respectively, 3.486 ± 0.145, 4.574 ± 0.222 and 6.608 ± 0.366; while the control group 99mTc- HYNIC-siRNA and 99mTcO4- group after injection of 1h, 4h and 10h when tumor imaging are unclear; control group, 99mTc- HYNIC-siRNA, 1h, 4h , 10h T/M ratio were 1.286 ± 0.189, 1.348 ± 0.204 and 1.354 ± 0.238; 99mTcO4- group 1h, 4h, 10h T/M ratio were 1.317 ± 0.164, 1.322 ± 0.159 and 1.401 ± 0.145. In each imaging time point, three groups of T/M ratio difference was statistically significant (F1 = 570.2, F4 = 903.1, F10 = 1294, P <0.01). In the T/M ratio for each time point imaging interference group were significantly higher than the free 99mTcO4- and the control group, the difference was statistically significant (t control = 29.20, 33.84 and 38.07, t free = 31.29, 37.61 and 41.86, P <0.01); the difference between the control group 99mTc- HYNIC-siRNA of T/M ratio and free 99mTcO4- group was not statistically significant (t = 0.392, 0.318 and 0.533, all P> 0.05). Imaging of tumor-bearing mice is shown in Figure 2.

Established breast cancer (MDA-MB-231) in tumor models
Gene imaging
Discussion: Study confirmed that tumor metastasis is organ-specific metastasis of tumor cells due to the high expression of CXCR4 expression site CXCL12, CXCL12 in the tumor cell chemotaxis specific transfer occurs, such as Muller et al. [22] in 2001 for the first time in Nature reported malignant breast primary tumor expression of chemokine receptor CXCR4, while the common sites of metastases ligand CXCL12 expression such as the lungs, bones and liver. Since then a large number of studies show that at least 23 different types of cancer cells the expression of CXCR4 [23], such as breast cancer, ovarian cancer, prostate cancer, colorectal cancer and malignant melanoma [22], and the expression of CXCR4 and closely related to prognosis. After CXCL12 CXCR4 can specifically bind to form molecules coupled through activation of downstream signaling pathways exert different biological effectiveness of different, resulting in embryonic development, immunity, inflammation, blood, HIV infection, angiogenesis, tumor invasion and metastasis a variety of physiological processes play a role in disease [22] (Figure 3).
Tumor gene expression imaging is the use of radiolabeled molecular probes, at the level of mRNA or protein in vivo noninvasively detect tumor specific gene product expression method of a developing situation, including antisense imaging, transfer gene expression and tumor imaging guide MDR gene expression imaging and the like. Developing small interfering RNA is a small interfering RNA labeled with radionuclide scintigraphy as a tracer to complementary base pairing based on the principle that small interfering RNA that specifically binds to the gene in the overall level dynamic monitoring for diseased tissue specific gene expression to achieve the genetic level in early qualitative diagnostic purposes disease. In previous imaging studies, tumor development in general will play a major role in oncogene radionuclide labeled probes made, but only for the corresponding probe a tumor, more limited application. Studies have shown that, CXCR4 in a variety of different types of cancer cells, overexpression, and in human breast cancer MDA-MB-231 cells over-expressed CXCR4 therefore chosen as target genes in human breast cancer gene imaging. The experiments selected for its consistent CXCR4 CXCR4-370 small interfering RNA, the sense strand sequence 5'GGGACUAUGACUCCAUGAATT3', antisense strand sequence 5'UUCAUGGAGUCAUAGUCCCTT 3 '.
In imaging studies, though by small interfering RNA backbone modifications (such as phosphorothioate modifications and borane phosphate modification), 2 'ribose modifications and other chemical modifications after nuclease resistance increased significantly, but we did not use. In this study, we used the 5 'end 6 carbon hexyl primary amine and ammonia structure, 2-deoxy-TdR d (TT) structure to the 3' end and full 2'-methoxy modified. After 2'-OMe modified small interfering RNA for a variety of physical and chemical factors of resistance has been greatly improved while the metabolic degradation of a variety of enzymes has also been some resistance, but also RNA molecules in vivo transport capacity also increased. In this experiment, 5 antisense strand 'end a six-carbon-hexyl primary amine and ammonia structure, as HYNIC coupled with a group of the 3' end two used to improve the antisense nucleotide binding activity deoxy thymus d (TT) structure. Full 2'-methoxy modified [23].
HYNIC and small interfering RNA chelation process, the excess of the small interfering RNA HYNIC ensure fully chelated HYNIC, therefore after chelation mixture containing mainly HYNIC-siRNA and free HYNIC. Using Sep-Pak C18 cartridges separation and purification, different substances due to different polarities and with the increasing concentration of the eluent is eluted successively. After coupling small interfering RNA purified by Sep-Pak C18 column and collected 80 effluents by UV spectrophotometer, the emergence of a thin high-260nm UV absorption peak at around 31, because the mixture does not contain free the siRNA, and therefore, this peak is produced by HYNIC-siRNA.
After using 99mTc HYNIC-siRNA interference group and control group HYNIC-siRNA mark, was measured using a spectrophotometer, visible both 260nm UV absorption peak, and the peak of the range of different sizes, according to Sephadex G25 chromatography column according to the separation of substances The principle determined by the relatively large peak molecular weight is relatively large 99mTc labeled successful HYNIC-siRNA generated by a relatively small peak molecular weight of a relatively small unmarked on 99mTc the HYNIC-siRNA production. Radiometric found only collect a wide range of peak tube containing radioactive liquid; also it proved the above conclusion.
In gene imaging, the probe can remain stable for a period of time is the key to determine the success of imaging, this experiment showed that small interfering RNA 99mTc labeled after PBS and incubated in fresh serum, a radioactive peak was See drift, indicating 120min markers within unseparated degradation while maintaining stability. Gene imaging, the purity level of the probe is to determine another key factor in the level of imaging quality, because labeling efficiency and radiochemical purity is too low, it may not work well probes specifically, if too much free technetium, bound to be thyroid tissue uptake, thyroid development, so that the probe imaging is poor. The results suggest that small interfering RNA after 99mTc mark Xinhua One use paper to paper chromatography, the stationary phase of acetone and saline as a double agent, measured application segment measurements obtained product labeling yield and radiochemical purity, the obtained results show that the product labeling efficiency and radiochemical purity are high.
In the living tissue complex physiological environment, small interfering RNA probe synthesis in tumor tissue is able to gather specific relation to the success of antisense imaging. Fu Peng [24] with the liposome-encapsulated DNA probe study probes nude mice bearing breast distribution, the results show that the oligonucleotides in nude mice of major organs such as liver, kidney excretion of radioactivity compared fast, but slow tumor area radioactivity cleared, tumor/ blood and tumor / skeletal muscle radioactivity ratio continues to rise over time, and at various points in time, the tumor to blood and tumor skeletal muscle liposome interference group probe set Radioactive counts were significantly higher than the ratio of liposome- encapsulated control probe group and 99mTcO4- group, thus confirming the probe synthesis can accumulate in tumor cells. Nimmagadda S et al [25 ] using radiopharmaceuticals labeled mouse anti-human CXCR4 monoclonal antibody in animal models, using SPECT/CT expression level of mouse body glioblastoma models in vivo CXCR4 were detected. The results showed that the site of tumor growth can be detected radiopharmaceuticals labeled monoclonal mouse anti-human CXCR4 antibody distribution. After, Nimmagadda et al. [25] and the application of Cu complexes CXCR4 antagonist AMD3100 as imaging agents, the use of PET for CXCR4 expression within the tumor were developing. Hartimath et al. [26] then use [99mTc] 02-AMD3100 CXCR4 receptor as a SPECT imaging tracers and successful conduct of the imaging.
SPECT imaging of nude mice bearing breast cancer showed that: interference group probes liposome-encapsulated within tumor lesions can gather specific and clear with time gradually, T/M ratio increased, while the control group in tumor probe intralesional radioactive nuclide sparse, free 99mTcO4- group, showing that a large number of radionuclide uptake from the image nude thyroid tissue, and tumor radioactive sparse, and interference probe set compared to further illustrate the tumor site gathered for the interference caused by the specificity of the probe, instead of radionuclides due to nonspecific aggregation. In summary, HYNIC indirect labeling of small interfering RNA ideal bifunctional chelator can be successfully used to mark the CXCR4 small interfering RNA probes. Radionuclide labeled probe not only remain stable in vitro, and small interfering RNA molecule probes by liposome after breast cancer cells can be gathered in and with the sequence of the target gene based on the principle of complementary base pairing specificity binding, with the necessary conditions for gene imaging. Small interfering RNA molecular probes capable of uptake in nude mice bearing breast cancer-specific area, the probe has to become a sensitive tracer of breast cancer diagnosis.
Conclusion
This oncogene is subject to CXCR4 intervention point to HYNIC bifunctional chelator, application of radionuclide 99mTc labeled CXCR4 mRNA in siRNA, marked rate, radiochemical purity, stability in vitro were analyzed. After and 99mTc-HYNIC siRNA injection of human breast cancer in nude mice in vivo SPECT imaging study performed for the synthesized 99mTc- HYNIC-siRNA enters whether specifically bind to a target gene in vivo mouse model of human breast cancer after for verification.
The results are as follows: to HYNIC as a chelating agent, can successfully labeled CXCR4, for small interfering RNA, in vitro labeled product can remain stable for some time, suitable for in vitro studies and in vivo imaging requirements. Imaging studies have shown that small interfering RNA 99mTc-labeled molecular probes of CXCR4 in human breast cancer nude mouse model of tumor tissue-specific aggregation, tumor tissue lesions can be displayed at the molecular level, as in the early non-invasive human breast cancer genetic diagnosis to provide a reliable theoretical basis.
References
- Foster CS, Vitale A (2012) Diagnosis and Treatment of Uveitis Second Edition. Jaypee Brothers Medical Publishers, New Delhi, India.
- Intraocular Inflammation and Uveitis. 2013-2014 Basic and Clinical Science Course. AAO, San Francisco, USA.
- Biswas J, Narain S, Das D, Ganesh SK (1996) Pattern of uveitis in referral uveitis clinic in India. IntOphthalmol 20(4): 223-228.
- De Smet M, Okada A (2010) Cystoid macular edema in uveitis. Dev Ophthalmol Basel, Karger,47: 136-147.
- Islam SM, Tabbara KF (2002) Causes of uveitis at The Eye Center in Saudi Arabia: A retrospectivereview. Ophthal Epidemiol 9(4): 239249.
- Suhler EB, Lloyd MJ, Choi D, Rosenbaum JT, Austin DF (2008) Incidence and prevalence of uveitis in Veterans Affairs Medical Centers of the Pacific Northwest. Am J Ophthalmol 146(6): 890-896.
- Nussenblatt RB, Whitcup SC, Uveitis (2010) Fundamentals and Clinical Practice Philadelphia, Elsevier, Netherlands.
- Prete M, Guerriero S, Dammacco R, Fatone MC, Vacca A, et al. (2014) Autoimmune uveitis: a retrospective analysis of 104 patients from a tertiary reference center. J Ophthalmic Inflamm Infect 24(4): 17.
- Rothova A (2007) Inflammatory cystoid macular edema. Curr Opin Ophthalmol 18(6): 487-492.
- Sallam A, Sheth HG, Habot-Wilner Z (2009) Outcome of raised intraocular pressure in uveitic eyes with and without a corticosteroid-induced hypertensive response. Am J Ophthalmol 148(2): 207-213.
- Sonam AB, Kumar V, Raina U, Ghosh B, Thakar M (2011) Inflammatory glaucoma. Oman J Ophthalmol 4(1): 3-9.






























