In Silico Optimization of Formate Formation by Acetobacteriumium woodii's formate Dehydrogenase
Bijan Bambaei1, Jafar Hemmat2, Ali Asghar Karkhane1 and Ibrahim Torktaz1*
1National institute of genetic engineering and biotechnology, Iran
2Biotechnology department, Iranian research organization for science and technology, Iran
Submission: December 18, 2016; Published: Junary 02, 2017
*Corresponding author: Ibrahim Torktaz, National institute of genetic engineering and biotechnology, Tehran, Iran, Email: torktaz@ut.ac.ir
How to cite this article: Bijan B, Jafar H, Ali Asghar K, Ibrahim T. In Silico Optimization of Formate Formation by Acetobacteriumium woodii's formate Dehydrogenase. Int J cell Sci & mol biol. 2017; 1(3): 555564. DOI:10.19080/IJCSMB.2017.01.555564
Abstract
Formate dehydrogenase from Acetobacteriumium woodii is under investigation for industrial conversion of CO2 to formate. To have an economically viable biotechnological process for production of formate from CO2 the conversion rate needs to be increased substantially. Here, we used structure prediction and molecular modelingfor predicting a reliable 3D structure of A. woodii FDH and following docking experiments we identified two critical residues as main formate binding sites in the enzyme. Our result will help to increase the efficacy of CO2 fixation as a biotechnological approach to reduce the atmospheric concentration of this greenhouse gas as well as biologically producing formate as a safe way of H2 transport in biofuel application.
Keywords: Formate; Acetobacteriumium woodii; Formate dehydrogenase; Docking
Introduction
In September 2016 the concentration of atmospheric CO2 has reached to 400 ppm (NASA). The increased in CO2, alongside other greenhouse gases has profound effects in climate all over the world and threatens a sustainable life on the planet in long run. The reduction of atmospheric CO2 is an interesting challenge for chemists as well as biochemists [1,2]. The biotechnological conversion of CO2 to organic acids via enzymatic processes other than photosynthesis is an attractive alternative. One of these enzymatic solutions is conversion of CO2 to formate catalyzed by the action of formate dehydrogenase (FDH; E.C.:1.2.1.2).
Although most known FDHs predominantly convert formate to CO2, in acetogenic bacteria the tendency is toward reduction of CO2 to formate [3]. The eminence of developing new biocatalysts with favorable kinetics towards formate production has motivated research in this area (Hartmann). The FDH of Acetobacteriumiumwoodii is one of the few characterized enzymes for this purpose. A. woodii is now being considered as a key player in biotechnological conversion of CO2 and H2 to formate for safe transport of hydrogen gas [4]. Unfortunately, the crystal structure of this enzyme is not publicly available. Therefore,any attempt for improving the kinetics of this enzyme demands the prediction of 3 dimensional structureof the enzyme based on homology with other FDH's. Here, we sought to increase formate production by A. woodii's FDH by in silico modelling, molecular docking and simulated site-directed mutagenesis to enhance formate release from the active site.
Methodology
The ligand structures were obtained from Pubchem database (molybdopterin: CID_45266731 and formate: CID_283) and the sequence of formate dehydrogenase from Acetobacteriumiumwoodii was obtained from NCB database (accession # WP_041668236.1).The 3D structure of protein was predicted by a Meta prediction approach. To do this, we have used phyre2, a fold recognition algorithm to predict the model based on the identical folds from known homologues structures [5]. In addition, we have used I-tasser, an ab inotio based prediction web server [6]. The obtained structures were used as the template for final model prediction by Modeller V 9.12 software [7]. The predicted structure was verified by Prosa Z score [8] and solved in a water box including neutralizing ions using GROMACS 5.0 software [9]. The solved structure was used as the template for molybdopterin interaction prediction site by docking method using mole gro virtual docker software [10] by MolDock as scoring function. After docking energy minimization performed and hydrogen bonds optimized to reach highly possible accurate binding model.
Results and Discussion
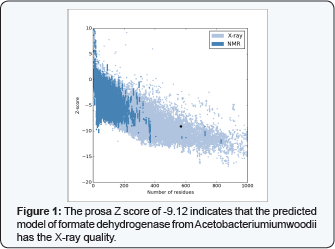
Our approach for enhancing formate formation is based on decreasing the binding affinity of formatein the active site of the formate dehydrogenase enzyme, the catalytic equilibrium would be redirected to formate release and hence providing space for new CO2 and H2 substrate molecules. The critical step was to predict a reliable 3D structure. The predicted structure reached the score of -9.12 in Prosa Z scoring system which means that the predicted model has the X-ray quality. The quality of predicted model form described Meta prediction method is depicted in (Figure 1) (Table 1). Formation from CO2 and H2, so it would be a better candidate for being used in the formic acid production industry. For gaining this purpose, we attempt to reach a high quality theoretical model.
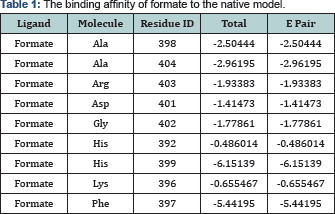
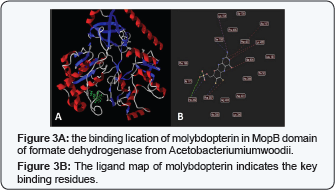
The primary structure comparison in the conserved Molybdopterin-Binding (MopB) domain of the MopB superfamily of proteins, a large, diverse, heterogeneous super family of enzymes that, in general, bind molybdopterin as a cofactor is present in (Figure 2) (Table 2). This domain is the first 413 N-terminal residues of the enzyme. To investigate which amino acids are playing key role in ligand binding, we have docked molybdopterin to the Mop B domain of the enzyme. Molybdopterin is present in the catalytic site of enzyme and facilitate the electron transfer. The interaction of top 20 hits with Molybdopterin-Binding domain was investigated and observed that most of the hits were located in one position of the Mop B, so with the most probability this position is the most possible site for ligand binding. The ligand map and the location of binding position is depicted in (Figure 3) (Table 3). Also the structure of formate was docked in the predicted active site position in the presence of Molybdoptein. The ligand map and the position of formate in the active site is depicted in (Figure 4.) Based on the ligand map, it can be understood that residues F397 and H399 are key amino acids in formate binding while they are not directly engaged in the binding with molybdopterin. So, performing substitution in these positions are favorable for decreasing the binding affinity of formate to the active site. For gaining this purpose, we have designed several FDH mutant models by replacing F397 and H399 with different type residues and similar in steric condense.

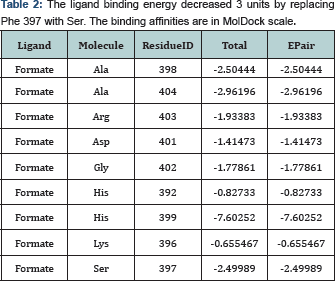
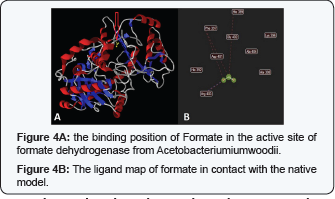
We have selected residues with similar steric condense to apply the minimum possible change in the conformation of protein and avoid alteration in the secondary structure pattern of the substitution location. Phe397 made hydrophobic interaction with the carbon atom of formate. For debilitation of Phe 397- C1 binding, we have replaced Phe 397 with Serine and the results of docking indicated that the binding affinity decreased significantly (3 units in Mol Dock scale). Also His 399 made a steric interaction with C1 from formate. We have replaced it with Methionine and found that the binding affinity decreased 6.1 unit in molDock scale.
Docking simulation of formate revealed new critical residues as targets for protein engineering experiments. Previous attempts for improving FDHs efficacy were concentrated on NAD binding residues [11], or cofactor regeneration for making a bifunctional enzyme [12]. Also the catalytic efficiency of formate dehydrogenase from Candida boidinii was increased in a study before [13]. Moreover, by site saturation mutagenesis the catalytic activity of Candida methylica formate Dehydrogenase was increased [14]. Therefore, our approach opens new area for protein engineering of FDH with aim of increasing the formate formation capacity of A. woodii’s FDH.
Conclusion
Macromolecular simulation of formate Dehydrogenase from Acetobacteriumiumwoodii indicated that Phe 397 and His 399 are key amino acids which bind to the catalytic product formate. Replacing Phe 397 with Ser and His 399 by Met theoretically will decrease the avidity of formate and makes better condition for its release from the active site.
References
- Taheri A, Berben LA (2016) Making C-H bonds with CO2: production of formate by molecular electrocatalysts. Chem Commun (Camb) 52(9): 1768-1777.
- Shi J, Jiang Y, Jiang Z, Wang X, Wang X, et al. (2015) Enzymatic conversion of carbon dioxide. Chem Soc Rev 44(17): 5981-6000.
- Alissandratos A, Kim HK, Easton CJ (2013) Formate production through biocatalysis. Bioengineered 4(5): 348-350.
- Schuchmann K, Muller V (2013) Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342(6164): 1382-1385.
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015 ) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6): 845-858.
- Yang J, Yan R, Roy A, Xu D, Poisson J, et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1): 7-8.
- Webb B, Sali A (2014) Protein structure modeling with MODELLER. Methods Mol Biol 1137: 1-15.
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(Web Server issue): W407-W4010.
- Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7): 845-854.
- Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49(11): 3315-3321.
- Hoelsch K, Suhrer I, Heusel M, Weuster-Botz D (2013) Engineering of formate dehydrogenase: synergistic effect of mutations affecting cofactor specificity and chemical stability. Appl Microbiol Biotechnol 97(6): 2473-2481.
- Jiang W, Fang BS (2016) Construction and evaluation of a novel bifunctional phenylalanine-formate dehydrogenase fusion protein for bienzyme system with cofactor regeneration. J Ind Microbiol Biotechnol 43(5): 577-584.
- Jiang W, Lin P, Yang R, Fang B (2016) Identification of catalysis, substrate, and coenzyme binding sites and improvement catalytic efficiency of formate dehydrogenase from Candida boidinii. Appl Microbiol Biotechnol 100(19): 8425-8437.
- Ozgun GP, Ordu EB, Tutuncu HE, Yelboga E, Sessions RB, et al. (2016) Site Saturation Mutagenesis Applications on Candida methylica Formate Dehydrogenase. Scientifica (Cairo) 2016: 4902450.






























