Prediction of Novel MicroRNA Located in Forkhead Box R2 Gene
Sara Mohammadi1, Maryam Hassanlou1* and Amir-Reza Javanmard2*
1Farzanegan campus, Semnan University, Iran
2Department of Molecular Genetics, Faculty of Biological Sciences, Tarbiat Modares University, Iran
Submission: May 05, 2023; Published:June 06, 2023
*Corresponding author: Maryam Hassanlou, Farzanegan campus, Semnan University, Semnan, Iran Email: Amir-Reza Javanmard, Department of Molecular Genetics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran, Email: amirjavan1372@gmail.com
How to cite this article: Sara M, Maryam H, Amir-Reza J. Prediction of Novel MicroRNA Located in Forkhead Box R2 Gene. Glob J Reprod Med. 2023; 10(1):8055779. DOI: 10.19080/GJORM.2023.10.555781.
Abstract
Forkhead Box R2 (FOXR2) is a transcription factor located on the X chromosome and its transcription normally restricted to the testis. FOXR2 is aberrantly expressed in 70% of all cancers. FOXR2 promotes formation of CNS-embryonal tumors, pediatric brain cancers and subsets of medullo-, pineo- and glioblastoma in CNS. MicroRNAs (miRNAs) are endogenous, small non-coding RNAs that function in regulation of gene expression. Compelling evidence have demonstrated that miRNA expression is dysregulated in human cancer through various mechanisms, including amplification or deletion of miRNA genes, abnormal transcriptional control of miRNAs, dysregulated epigenetic changes and defects in the miRNA biogenesis machinery. MiRNAs may function as either oncogenes or tumor suppressors under certain conditions. The dysregulated miRNAs have been shown to affect the hallmarks of cancer, including sustaining proliferative signaling, evading growth suppressors, resisting cell death, activating invasion and metastasis, and inducing angiogenesis. An increasing number of studies have identified miRNAs as potential biomarkers for human cancer diagnosis, prognosis and therapeutic targets or tools, which need further investigation and validation. In this research, new microRNAs in FOXR2 gene were predicted using bioinformatics tools. SSC profiler program was utilized to predict the stem-loop structures within the genomic area of FOXR2 gene. UCSC genome browser database was used to analyze the conservation status of the putative miRNA and its precursor sequence. Furthermore, the FOXR2-miRNA’s mature sequence was also performed by MatureBayes online tool. In addition, RNAFOLD online software which applies the minimum-free energy (MFE) RNA structure prediction algorithm, was used for approximate prediction of the stem-loop secondary structure. Our results demonstrate that FOXR2 with about 1925 bp length has one miRNA stem-loop-like structure that has relatively conserved sequences. Overall, accumulative pieces of evidence indicated the presence of a novel miRNA encoded within the FOXR2.
Keywords: Cancer; Testis and transcription factor; FOXR2
Introduction
FOX proteins are a superfamily of evolutionarily highly conserved transcriptional regulators (TFs) that all share a fork-like DNA binding domain [1]. These TFs play a wide role in cellular homeostasis and their expression is well controlled in mature tissues. In this large family of TFs, FOXR2, located on the X chromosome, has been implicated as an oncogene in a subset of cancers including reproductive tracts [2-6]. However, there is still no systematic characterization of the oncogenic role of FOXR2 in all cancers, and the mechanisms through which it induces tumorigenesis are not fully elucidated. Although FOXR2 has been shown to be activated through constitutive variants in the central nervous system [7], and peripheral neuroblastomas [5], the expression profile of FOXR2 in all cancers and the different genetic and epigenetic mechanisms of FOXR2 expression activation remain to be elucidated. Furthermore, the pattern and regulation of FOXR2 expression in normal tissues is unknown. The mechanisms through which FOXR2 promotes tumorigenesis have also not been systematically evaluated. TFs usually exert their effects by cooperating in transcriptional complexes. Indeed, some TFs, including other members of the junction box family, have been shown to mediate their effects by dimerizing directly with other TFs or by associating with distinct DNA regions [8-12]. Analysis of FOXR2 expression among a cohort of human cancers that includes both adult [13,14], and pediatric data sets shows evidence of FOXR2 expression in more than 70% of cancer types and distinct mechanisms of FOXR2 activation, including a novel nonstructural mechanism has been shown to account for most FOXR2-expressing cancers [15-18]. These findings highlight the unrecognized role of FOXR2 as a potent oncogene in human cancers and show how TFs from different families cooperate to induce oncogenes. In this research, considering the complex function of FOXR2 gene, we predicted the presence of miRNAs in this gene. It is possible to consider part of the activities of this gene to be related to the miRNAs that are inside this gene.
Methods
Bioinformatic prediction of miRNA presence in FOXR2 gene
To predict the presence of miRNA stem-loop structures, SSCprofiler site at http://miRNA.imbb.forth.gr/SSCprofiler. html was used. This site indicates the possibility of miRNA-like secondary structures in the target sequence. This web portal suggests the hairpin structures and mature sequence of a miRNA based on features such as sequence, structure and conservation. From the point of view of this software, in the secondary structures of stem-loop similar to miRNA, the number of protrusions should be less than 16 and the number of loops should be less than 32, the average number of loops plus protrusions should be less than 37 and more than 25% of nucleotides must be protected. The minimum free energy defined for that structure must be less than -25.44kcal/mol. In this software, the score threshold is 3, and the higher the score, the more likely the prediction is correct. For this purpose, the genomic location of the desired gene is obtained as a number through hg17 available on the UCSC website. Each time, the sequence of 1000 base pairs is scanned in both positive and negative strands, and in order to cover the marginal regions of the sequence, the sequences are scanned once from the beginning of the sequence number obtained on the UCSC site plus 500 until the marginal regions be covered too.
Prediction of the secondary structure (stem-loop) of the miRNA sequence
For this purpose, use the RNAfold website at http://rna. tbi.univie.ac.at/http://rna.tbi.univie.ac.at/ became. RNAfold calculates and maps the minimum free energy (MFE) structure of given sequences. If the minimum free energy of the series pin structure is less than -25.44 kcal/mol, then the structure has good stability. The structure is colored by base pairing probabilities. The probability range is from 0 to 1. 1 is the highest value and is marked with red color and 0 is the lowest value and is marked with blue color.
Evaluation of miRNA sequence conservation: To analyze the degree of conservation of predicted miRNA sequences, the BLAT section of the UCSC site at http://genome.ucsc.edu/ was used. On this website, sequence conservation in 17 different species is shown in green. Prediction of mature miRNA sequence: The MatureBayes site at http://mirna.imbb.forth.gr/MatureBayes.html was used to find the mature sequence of miRNAs. MatureBayes provides the most likely start position of the mature miRNA(s) in each miRNA precursor.
Results
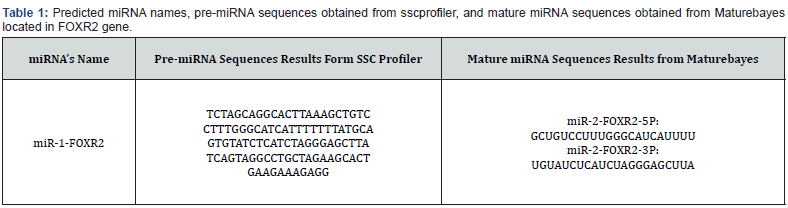
Bioinformatic prediction of the presence of miRNA in FOXR2 gene: With the investigations carried out on the SSCprofiler site, a miRNA was identified in exon 1 of the FOXR2 gene (Figure 1), whose precursor sequence is listed in table 1. Also, the mature miRNA sequence of these miRNAs was predicted with the help of the MatureBayes site (Table 1). The location of miRNA precursor sequence obtained in the exonic and intronic regions of the gene was obtained with the help of ensemble site (https://asia. ensembl.org/index.html).
Predicting miRNA stem-loop structure and checking its degree of conservation: With the help of the RNAfold online website, the stem-loop structure (Stem-loop) of the identified miRNAs was predicted (Figure 2a). Analysis of the pre-miRNA sequence using the UCSC website showed that this sequence lacked conservation among 17 different species (Figure 2b).
Conclusion
So far, no microRNA has been reported in the FOXR2 gene. During the investigations, it was found that there is a miRNA stemloop sequence in the FOXR2 gene, which has not been reported so far. To predict the presence of microRNA, we first used the SSCprofiler site, then we predicted the secondary structures of microRNA with the help of the RNAfold site. We assessed sequence conservation using the UCSC site. To predict the mature structure of microRNAs, the site Mature Bayes was used, which showed us the most probable mature structures of microRNAs. So far, bioinformatics methods have been used in several studies to identify new miRNAs. The method used in this study has previously been used for the bioinformatic identification of miRNAs, whose existence has been confirmed after prediction using experimental methods [19-21]. Therefore, it seems that bioinformatics methods can help identify new miRNAs in the human genome.


References
- Myatt SS, EW Lam (2007) The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 7(11): 847-859.
- Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, et al. (2013) Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 45(7): 756-766.
- Koso H, Tsuhako A, Lyons E, Ward JM, Rust AG, et al. (2014) Identification of FoxR2 as an oncogene in medulloblastoma. Cancer research 74(8): 2351-2361.
- Beckmann PJ, Larson JD, Larsson AT, Jason P Ostergaard, Sandra Wagner, et al. (2019) Sleeping Beauty Insertional Mutagenesis Reveals Important Genetic Drivers of Central Nervous System Embryonal Tumors. Cancer Res 79(5): 905-917.
- Braoudaki M, Kyriaki Hatziagapiou, Apostolos Zaravinos, George I Lambrou (2021) MYCN in Neuroblastoma: "Old Wine into New Wineskins". Diseases 9(4): 78.
- Zhang S, Jie Cheng, Chenlian Quan, Hao Wen, Zheng Feng, et al. (2020) circCELSR1 (hsa_circ_0063809) Contributes to Paclitaxel Resistance of Ovarian Cancer Cells by Regulating FOXR2 Expression via miR-1252. Mol Ther Nucleic Acids 19: 718-730.
- Sturm D, Brent A Orr, Umut H Toprak, Volker Hovestadt, David TW Jones, et al. (2016) New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164(5): 1060-1072.
- Cirillo L, Frank Robert Lin, Isabel Cuesta, Dara Friedman, Michal Jarnik, et al. (2002) Opening of Compacted Chromatin by Early Developmental Transcription Factors HNF3 (FoxA) and GATA-4. Molecular cell 9: 279-289.
- Seo S, Hideo Fujita, Atsushi Nakano, Myengmo Kang, Antonio Duarte, et al. (2006) The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol 294(2): 458-470.
- Shu W, Min Min Lu, Yuzhen Zhang, Philip W Tucker, Deying Zhou, et al. (2007) Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134(10): 1991-2000.
- Bass AJ, Hideo Watanabe, Craig H Mermel, Soyoung Yu, Sven Perner, et al. (2009) SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41(11): 1238-1242.
- Kume T (2009) The cooperative roles of Foxc1 and Foxc2 in cardiovascular development. Adv Exp Med Biol 665: 63-77.
- Weinstein JN, Eric A Collisson, Gordon B Mills, Kenna R Mills Shaw, Brad A Ozenberger, et al. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45(10): 1113-1120.
- Barretina J, Giordano Caponigro, Nicolas Stransky, Kavitha Venkatesan, Adam A Margolin, et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391): 603-607.
- Dubois FPB, Ofer Shapira, Noah F Greenwald, Travis Zack, Jeremiah Wala, et al. (2022) Structural variants shape driver combinations and outcomes in pediatric high-grade glioma. Nat Cancer 3(8): 994-1011.
- Van Tilburg CM, Elke Pfaff, Kristian W Pajtler, Karin PS Langenberg, Petra Fiesel, et al. (2021) The Pediatric Precision Oncology INFORM Registry: Clinical Outcome and Benefit for Patients with Very High-Evidence Targets. Cancer Discov 11(11): 2764-2779.
- McLeod C, Alexander M Gout, Xin Zhou , Andrew Thrasher, Delaram Rahbarinia, et al. (2021) St. Jude Cloud: A Pediatric Cancer Genomic Data-Sharing Ecosystem. Cancer Discovery 11: CD-20.
- Worst BC, Cornelis M van Tilburg, Gnana Prakash Balasubramanian, Petra Fiesel, Ruth Witt, et al. (2016) Next-generation personalised medicine for high-risk paediatric cancer patients - The INFORM pilot study. Eur J Cancer 65: 91-101.
- Rahimi A, Rina Sedighi, Modjtaba Emadi-Baygi, Mohammad-Amin Honardoost, Seyed-Javad Mowla, et al. (2020) Bioinformatics prediction and experimental validation of a novel microRNA: hsa-miR-B43 within human CDH4 gene with a potential metastasis-related function in breast cancer. J Cell Biochem 121(2): 1307-1316.
- Dokanehiifard S, Bahram M Soltani, Sepideh Parsi, Fahimeh Hosseini, Mohammad Javan, et al. (2015) Experimental verification of a conserved intronic microRNA located in the human TrkC gene with a cell type-dependent apoptotic function. Cell Mol Life Sci 72(13): 2613-2625.
- Parsi S, Bahram M Soltani, Ebrahim Hosseini, Samaneh E Tousi, Seyed J Mowla (2012) Experimental verification of a predicted intronic microRNA in human NGFR gene with a potential pro-apoptotic function. PLoS One 7(4): e35561.






























