Autophagy-Related Genes mRNA Expression in Patients with Otitis Media with Effusion
Myung Gu Kim1, Su Young Jung2, Sung Su Kim3, Sung Ho Cha4, Yong Sung Choi4, Young Il Kim5, Sang Hoon Kim2 and Seung Geun Yeo2,5*
1Department of Otorhinolaryngology, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Republic of Korea
2Departments of Otorhinolaryngology, School of Medicine, Kyung Hee University, Korea
3Department of Biochemistry and Molecular Biology, Medical Science and Engineering Research Center for Bioreaction to Reactive Oxygen Species, School of Medicine, Kyung Hee University, Korea
4Department of Pediatrics, School of Medicine, Kyung Hee University, Korea
5Medical Science Research Institute, Kyung Hee University Medical Center, Korea
Submission: March 14, 2018; Published: April 02, 2018
*Corresponding author: Seung Geun Yeo, Department of Otorhinolaryngology Head and Neck Surgery, Kyung Hee University, Korea;Tel: 82-2-958-8474; E-mail:yeo2park@gmail.com
How to cite this article: Myung G K, Su Young J, Sung S K, Sung H C, Yong S C, et al. Autophagy-Related Genes mRNA Expression in Patients with Otitis Media with Effusion. Glob J Oto 2018; 14(2): 555882. DOI: 10.19080/GJO.2018.14.555882
Abstract
Objective: Bacterial infection is a major cause of otitis media with effusion (OME) in patients with dysfunctional Eustachian tubes. Immune reactions to bacterial infection in the middle ear cavity are associated with chronic OME. This study evaluated the expression of autophagy- related genes mRNAs in effusion fluid.
Methods: Effusion fluid samples were obtained at the time of ventilation tube insertion in 97 patients diagnosed with OME. The levels of mRNA encoding autophagy-related genes in effusion fluid were measured by real-time RT-PCR. Levels were compared in patients with and without bacteria in effusion fluid, and relative to fluid characteristics.
Results: Autophagy-related genes mRNAs were present in all effusion fluid samples. Levels of autophagy-related genes mRNAs were lower in effusion fluid with than without bacteria, with mTOR and Bcl2 mRNA levels differing significantly (p<0.05 each). However, the levels of expression of autophagy-related genes mRNAs were unrelated to effusion fluid characteristics (p>0.05 each).
Conclusion: Autophagy-related genes mRNAs are involved in the pathophysiology of OME. The levels of mTOR and Bcl2 mRNAs were significantly lower in effusion fluid with than without bacteria.
Keywords: Oncology; Head and neck; Endoscopy; Training; Clinic
Keywords: Otitis Media with Effusion (Ome); Autophagy-Related Genes; m Tor; Bcl2 y
Introduction
Otitis media with effusion (OME) is characterized by the accumulation of fluid in the middle ear cavity without symptoms of acute infection. OME is frequently associated with bacterial infection of middle ear fluid in children with Eustachian tubes dysfunction [1]. Autophagy is a process during which substances in a cell surrounded by a double layer structure are passed on to lysosomes for degradation. Along with the ubiquity proteasome system, autophagy plays an important role in protein quality control in cells. In addition to its role in protein breakdown, autophagy is important in removing damaged organelles (e.g., mitochondria, endoplasmic reticulum, peroxisomes) and getting rid of extracellular pathogens (e.g. bacteria, viruses, parasites). In OME, autophagy may be involved in immune responses to bacterial infections in the middle ear. Deregulation of autophagy can cause various diseases, including cancers, degenerative brain diseases, infections, aging, Crohn's disease, and heart disease [2-5]. In the absence of nutrition, autophagy also maintains the energy balance in a cell, by generating ATP from the breakdown of proteins, lipid droplets and glycogen into amino acids, fatty acids, and glucose, respectively. This process, however, is inhibited under conditions of sufficient nutrition, allowing energy to be stored. Autophagy induced damage can result in an energy imbalance and may lead to other metabolic illnesses, including diabetes mellitus, obesity, and atherosclerosis [6-8].
Despite evidence showing the importance of autophagy in various metabolic disorders, most studies of the relationships between autophagy and disorders of energy metabolism and metabolic diseases have been performed in knockout mice without autophagy control genes. Therefore, it is necessary to evaluate autophagy in human subjects and in mice with various metabolic diseases and a dysfunctional autophagic process. Studies assessing the expression patterns of autophagy-related genes mRNAs in patients with OME, with and without bacterial infection of effusion fluid, are needed, as are studies analyzing the fluid characteristics on autophagy-related genes mRNAs. This study therefore assessed the effects of these factors on the levels of autophagy-related genes mRNA expression in effusion fluid samples from patients with OME.
Materials and Methods
Subjects
The study included 97 patients evaluated at Department of Otorhinolaryngology-Head and Neck Surgery at tertiary referral university hospital and diagnosed with OME. OME was diagnosed based on the amber color of effusion fluid, the airfluid level, air bubbles, or opacity of the tympanic membrane and B or C type tympanograms. Patients diagnosed with OME received medication for 1 or 2 weeks and were observed for 3 months. If no improvement was observed, patients underwent ventilation tube insertion, with effusion fluid samples obtained during surgery. Patients with retraction of the tympanic membrane or pure tone audio metry greater than 40 dB also underwent tube insertion, as did patients or patients whose parents desired the operation. The external auditory meatus was thoroughly disinfected without bleeding, and exudates were collected in an Eppendorf tube using a collector (Xomed Trace Products, Jacksonville, FL, USA). Samples were maintained at a temperature < -70 °C. The ear cavity was fully disinfected, and ventilator tubes were inserted by aseptic manipulation. Patients suspected of having head or neck anomalies, systemic diseases, or congenital or acquired Immuno deficiencies were excluded. The purpose of the study was explained to all patients and/or their parents or guardians, with all providing written informed consent for use of patient samples. The study protocol was approved by the institutional review board.
Bacterial Culture
Effusion fluid samples were obtained using sterile cotton swabs (Xomed Trace Products, Jacksonville, FL, USA) immersed in Stuart transport medium. Aliquots of these samples were added to dishes of solid blood agar and flasks containing liquid thioglycollate medium (Hangang, Kun-po, Korea). The cultures were incubated for 24h at 35 °C. Bacteria that formed colonies were identified by Gram staining and biochemical testing, and fungi successfully cultured in solid blood agar were subsequently grown in Sabouraud's medium.
RNA Extraction and Real-Time PCR
Total RNA was purified from homogenized tissue samples using TRIzol reagent according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). First-strand c DNA was synthesized from 1μg total RNA using a reverse transcription system with random hexamers (Promega, Madison, WI, USA), as described by the manufacturer. Real-time RT-PCR was performed on a StepOnePlus real-time PCR system. Each 20μl sample contained 1μl of c DNA 10μl Power SYBR Green PCR Master Mix (Applied Bio systems, Foster City, CA, and USA), 2μl of primers (Table 1), and 7μl of PCR-grade water. The amplification conditions consisted of an initial denaturation at 95°C for 10min, followed by 40 cycles of denaturation at 95°C for 15s and annealing and extension at 60°C for 1min. The crossing points of the target genes with p-actin were calculated using the formula
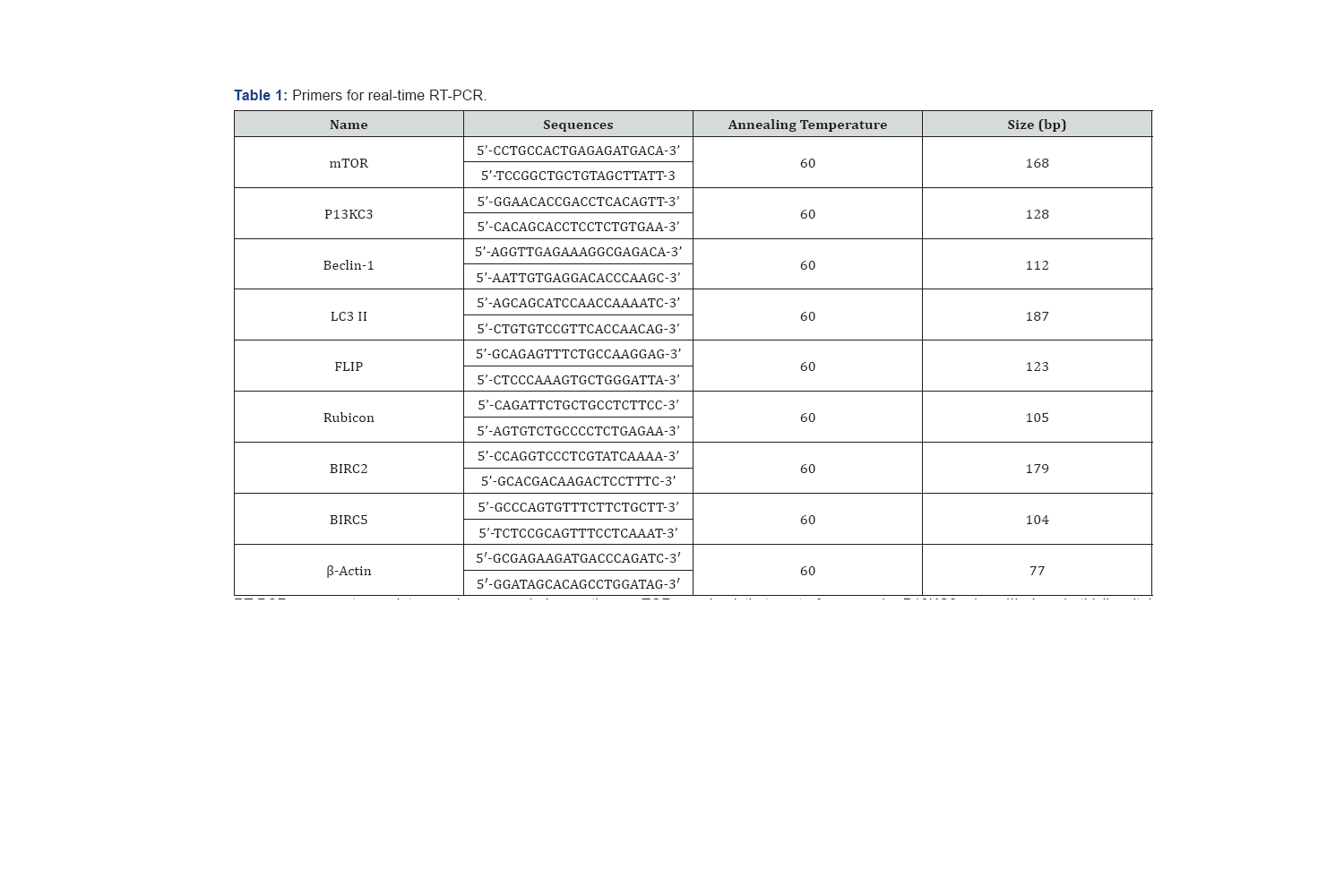
RT-PCR: reverse transcriptase-polymerase chain reaction, m TOR: mechanistic target of rapamycin, P13KC3: class III phosphatidylinositol 3-kinase, Bcl-2: B cell lymphoma 2, LC3 II: light chain 3-II, FLIP: FLICE-inhibitory protein, FLICE: FADD-like IL-1a-converting enzyme, BIRC: baculoviral IAP repeat-containing protein.
Statistical analysis
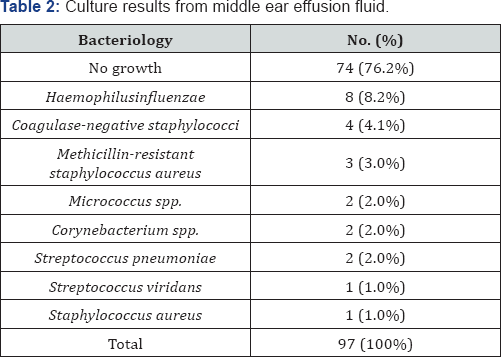
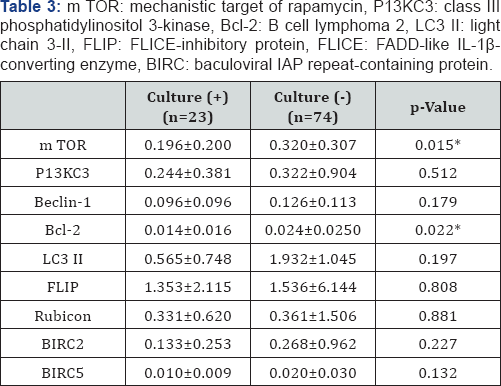
Data were analyzed statistically using IBM SPSS version 20 (SPSS Inc., Chicago, IL, USA) for Windows. Between groups comparisons were performed using Fisher's exact test and levels of mRNA were compared using the Levene homogeneity of variance test and Pearson’s correlation test. Statistical significance was set at a p value <0.05. The 97 patients with OME consisted of 53 males and 44 females, ranging in age from 0-80 years (mean age, 23.3 ± 26.5 years). Bacteria were detected in 22/97 (23.5%) effusion fluid samples by standard culture, with the remaining samples 75/97 (76.5%) being negative for bacterial growth. The most frequently detected bacterial species was Homophiles influenza, followed by coagulate negative Staphylococcus, MRSA (methicillin-resistant Staphylococcus aureus), Streptococcus pneumonia, Corynebacteriumspp,Micrococcus spp, Gram positive rods, Streptococcus viridans, and Gram negative rods (Table 2). Autophagy-related genes mRNAs were present in the effusion fluid of all 97 patients. The level of expression of autophagy-related genes mRNAs was higher in the culture negative than in the culture positive group. Specifically, mTOR and Bcl-2 mRNAs were significantly lower in effusion fluid from culture positive than culture negative patients p<0.05 each; (Table 3). However, the levels of autophagy-related genes mRNAs were unrelated to exudates characteristics p>0.05 (Table 4).
Discussion
Autophagy is an adaptive response that provides metabolites and is involved in tumor growth inhibition, cell removal of toxic substances, removal of microorganisms that invade cells, and antigen-presentation by macrophages and dendrite cells to lymphocytes [9]. Autophagy can be divided into several stages, including initiation, vesicle nucleation, vesicle elongation, fusion, degradation, and termination. This process is elaborately regulated by proteins produced during each step, with autophagy induction and inhibition involving several factors. This study investigated the expression patterns of several autophagy-related genes in patients with OME. Genes investigated included mTOR, which is involved in the induction and initiation of autophagy; P13KC3 and Beclin-1, which are involved in nucleation; BCL-2, which is an antagonist of Beclin-1; LC3 II, which is involved in auto phagosome biogenesis; FLIP, which is involved in vesicle elongation and is an antagonist of the autophagy-related gene (Atg); Rubicon, which inhibits maturation; and BIRC2 and BIRC5, which are antagonists to Rubicon [10-13].
The associations of autophagy with human health and human disease remain unclear. Although many studies have shown that autophagy has positive effects on some human diseases and negative effects on others, the mechanisms linking autophagy and disease are very complex. Activation of autophagy acts as a tumor suppressor, whereas inactivation of autophagy allows cancer cells to survive under nutrient-poor conditions. In muscular disorders, activation of autophagy may compensate for defects in lysosome function; whereas inactivation of autophagy may lead to the accumulation of autophagosomes, which may impair cell function. During the process of neurodegeneration, activation of autophagy allows the removal of protein aggregates before they become toxic, whereas inactivation of autophagy may induce cell death of neurons that accumulate aggregated proteins. Activation of autophagy by pathogens enhances cellular defenses against invasion by bacteria and viruses, whereas inactivation of autophagy allows pathogens to establish a replicative niche [14,15].
The prevalence of OME is high in patients with frequent upper respiratory infections (URI). Bacteria have been found in middle ear effusion of many of these patients, suggesting that bacterial infections may cause exudative otitis media [16]. Understanding bacterial infection and immune responses in the middle ear cavity may help determine the pathophysiology of OME [17]. Autophagy may be involved in the removal of pathogens that enter the cytoplasm through the cell membrane or through phagosomes,suggesting that bacterial infection may activate autophagy [18].This study therefore assessed whether autophagy associated associated with immune responses to bacterial infections is involved to the pathogenesis of OME. Autophagy-related genes were found to be expressed in exudates positive and negative for bacteria. The levels of expression were lower in exudates positive than negative for bacteria, perhaps because immune responses are impaired in the former. A normal, robust immune response to invading bacteria in the middle ear would include inactivation or eradication of invading bacteria, accompanied by increased expression of autophagy-related genes. In contrast, the presence of bacteria may indicate an abnormal or weak immune response, and a lower level of expression of autophagy-related genes. Similarly, inhibition of the activation of autophagy in adipose tissue was accompanied by increased expression of cytokines associated associated with the inflammatory response [19].
Antibiotic use early during the course of OM may eliminate bacterial infection of the middle ear, as well as affecting the expression of autophagy-related genes mRNAs. Nevertheless, the finding that autophagy-related genes are expressed in middle ear fluid both positive and negative for bacteria suggests that autophagy is involved in the pathogenesis of OME. Levels of expression of mTOR and Bcl-2 mRNAs were significantly lower in middle ear fluid positive than negative for bacteria. Because both mTOR and Bcl-2 are associated with inhibition of autophagy, these findings suggested that autophagy induction and inhibition occur simultaneously during the pathogenesis of OME. This study had several limitations. The negative culture rate was high, with only 23.2% of cultured middle ear fluid being positive for bacteria. This may have been due to the delayed collection of middle ear exudates from patients who developed OME after 3 months and the inefficiency of detection by conventional culture methods. Second, our findings may not fully reflect initial immune responses in the middle ear cavity because the subjects of this study were those with chronic OME. Third, although mRNAs were expressed, proteins may not have been. Fourth, we could not assess the expression of all genes involved in autophagy. Fifth, for ethical reasons, we could not harvest middle ear mucosa from the patients in this study and we therefore performed experiments on middle ear fluids. As these exudates contained only a few partially exfoliated mucosal epithelial cells and inflammatory cells, they could not fully reflect immune responses and inflammation in the middle ear mucosa during infection.
Conclusion
The present findings suggest that autophagy-related genes may be involved in the pathogenesis of OME. The levels of expression of mTOR and Bcl2 mRNAs were significantly lower in middle ear fluid positive than negative for bacteria.
Acknowle dgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2011-0030072).
References
- Jin Z, Cha SH, Choi YS, Kim YI, Shoi SA, et al. (2016) Expression of CXCL4 and aquaporin 3 and 10 mRNAs in patients with otitis media with effusion. Int J PediatrOtorhinolaryngol 81: 33-37.
- Kim KH, Lee MS (2014) Autophagy a key player in cellular and body metabolism. Nat Rev Endocrinol 10: 322-337.
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27-42.
- Mizushima N, Komatsu M (2008) Autophagy: renovation of cells and tissues. Cell 147: 728-741.?
- Choi AM, Ryter SW, Levine B (2013) Autophagy in human health and disease. N Engl J Med 368(7): 651-662.
- Singh R, Cuervo AM (2G11) Autophagy in the cellular energetic balance. Cell Metab 13: 495-5G4.
- Mizushima N (2007) Autophagy: process and function. Genes 21: 2861-2873.
- Russell RC, Yuan HX, Guan KL (2014) Autophagy regulation by nutrient signaling. Cell Res 24: 42-57.
- Zhang DX, Zhang JP, Hu JY, Huang YS (2G16) The potential regulatory roles of NAD (+) and its metabolism in autophagy. Metabolism 65: 4544621G. Yanaguchi O, Otsu K (2G12) Role of autophagy in aging. J Cardiovasc Pharmacol 60: 242-247.
- Yanaguchi O, Otsu K (2012) Role of autophagy in aging. J Cardiovasc Pharmacol 60: 242-247.
- Ghavami S, Shojaei S, Yeganeh B, Ande SR, Janqamreddy JT, et al. (2G14) Autophagy and apoptosis dysfunction in neurodegenerative disorders. ProgNeurobiol 112: 24-49.
- Mehrpour M, Esclatine A, Beau I, Codogno P (2G1G) Autophagy in health and disease 1 Regulation and significance of autophagy: an overview. Am J Physiol Cell Physio l298: C776-785.
- De Almagro MC, Vucic D (2012) The inhibitor of apoptosis (IAP) proteins are critical regulators of The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti cancer therapy. Exp On col 34: 200-211.
- Shntani T, Klionsky DJ (2004) Autophagy in health and disease; A double edged sword. Science 306: 990-995.
- Cuervo AM (2004) Autophagy; in sickness and in health. Trends in Cell Biol 14: 70-77.
- Lee HY, Kim YI, Lee JW, Byun JY, Park MS, et al. (2013) Decreased Expression of TLR 9 and Cytokines in the Presence of Bacteria in Patients with Otitis Media with Effusion. Clin Exp Otorhinolaryngol 6: 195-200.
- Lee SY, Ryu EW, Kim JB, Yeo SG (2011) Clinical approaches for understanding the expression levels of pattern recognition receptors in otitis media with effusion. Clin Exp Otorhinolaryngol 4: 163-167.
- Rich KA, Burkett C, Webster P (2003) Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 5: 455-468.
- Jansen HJ, Van Essen P, Koenen T, Joosten LA, Netea MG, et al. (2012) Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 153: 5866-5874.





























