Transcriptome Variation in Banded Newt (Ommatotriton Vittatus) During its Life Cycle and Adaptation to an Unpredictable Habitat
Gad Degani and Ari Meerson*
1MIGAL-Galilee Research Institute, P.O. Box 831, Kiryat Shmona 1101602, Israel
2Faculty of Sciences, Tel-Hai Academic College, Upper Galilee 1220800, Israel
Submission: November 09, 2023;Published: November 17, 2023
*Corresponding author: Ari Meerson, Faculty of Sciences, Tel-Hai Academic College, Upper Galilee 1220800, Israel. Email id: arim@migal.org.il
How to cite this article: Gad D, Ari M. Transcriptome Variation in Banded Newt (Ommatotriton Vittatus) During its Life Cycle and Adaptation to an Unpredictable Habitat Ecol Conserv Sci. 2023; 3(4): 555620. DOI:10.19080/ECOA.2023.03.555620
Abstract
Israel represents the southern border of the distribution of the banded newt (Ommatotriton vittatus). The life cycle of O. vittatus includes several distinct phases: eggs, aquatic larvae, a terrestrial phase and an aquatic reproductive phase. We investigated differences in gene expression during the life cycle and adaptations of banded newts to unpredictable (terrestrial and aquatic) habitats using mRNA-seq. We identified ~10k genes that were differentially expressed (DE) in one of the pairwise comparisons between 3 groups: 1 - terrestrial newts (males and females), 2 - aquatic newts (males and females), 3 - aquatic larvae before metamorphosis. The groups were clearly defined by Principal Components Analysis (PCA). The greatest difference was between aquatic newts (males and females) and aquatic larvae: ~7.4k DE genes. Of special interest were the ~2.4k genes DE between the aquatic and terrestrial phenotypes. These included prominent candidates with known roles in kidney function (uromodulin homologs were strongly associated with aquatic lifestyle), tissue structure (keratins), and the thyroid hormone signaling modulator DUOXA1. Additional developmental and metabolic pathways overrepresented among the identified DE genes included “epidermis development”, “nervous system development”, “nucleotide-sugar biosynthesis”. Overall, both metamorphosis and environmental adaptation of banded newts involve extensive transcriptomic remodeling involving developmental, metabolic, and cellular pathways. Understanding the roles of these pathways and individual genes is instrumental for studies of adaptation to unpredictable habitats, especially those affected by climate change. Furthermore, the phenotypic flexibility of the newt and the underlying regulation of gene expression can shed light on the evolution of terrestrial vertebrates.
Keywords: Banded Newt; Amphibians; Life Cycle; Metamorphosis; Environmental Adaptation; Transcriptome; Gene Expression; Mrna-Seq
Abbreviations: DE: Differentially Expressed; PCA: Principal Components Analysis; HCL: Hierarchical Clustering
Introduction
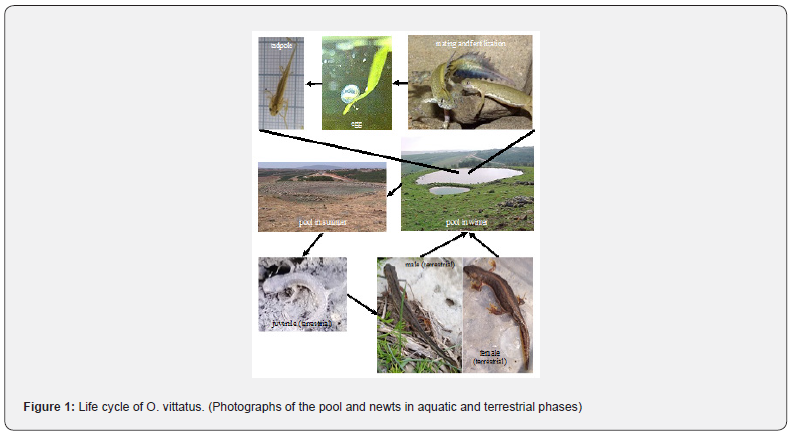
The banded newt (Ommatotriton vittatus, synonymous with Triturus vittatus) is one of three species of genus Ommatotriton (O. nesterovi, O. ophryticus and O. vittatus) found in Turkey, Syria, and Israel, and are adapted to extremely unstable conditions Degani [1]; Degani & Ahkked [2]; Van Riemsdijk [3] (Figure 1). The two species, O. ophryticus and O. vittatus, differ in trunk vertebra count, genome size and allozyme data. The northern taxon, O. ophryticus, is subdivided into two geographical fragments: the “western group” populations from western Anatolian Turkey, and the “eastern group” populations distributed in the rest of Turkey and Western Caucasus. O. vittatus is found in Israel Degani [1], which is the southern border of the genus’s distribution, and which is characterized by the most extreme seasonal changes in the environment. For example, the habitat described in our study is aquatic only ~1 month out of the year. This fact ostensibly makes the adaptability of the newt to the environmental changes especially significant for its survival, and represents a strong selective pressure for such adaptability, which is likely less pronounced in areas further north, where the climate is less arid. The adaptation of O. vittatus to the southern border of newt populations in Israel and many aspects of O. vittatus biology in Israel have been investigated: its life cycle Geffen [4]; Pearlson & Degani [5], ecological conditions during larval growth Degani [6]; Pearlson & Degani [5], genetic variation Degani [7] and environmental hiding-place seeking behavior after metamorphosis and genetic differentiation of the larvae in various breeding places Degani & Ahkked [2]. The development of the newt Triturus carnifex, which is very similar to O. vittatus, has been described from egg deposition to hatching and illustrated with the use of photographs of living embryos Damen [8]. The life cycle of O. vittatus includes several distinct phases: eggs, aquatic larvae, a terrestrial phase and an aquatic reproductive phase Degani [9] (Figure 1). Due to the extreme seasonal variation of its habitat, this species is thus particularly appropriate for a study of adaptation-related transcriptomic changes. In the current study, we compared the tail transcriptomes of aquatic larvae, terrestrial adults (females and males), and aquatic adults (females and males), in order to understand how changes in gene expression help this species adapt to its environment over the course of its life cycle.
Materials and Methods
Study Area
Nahalit Pool is a winter pool, located on the slopes of an agricultural settlement in the Upper Galilee mountains, among grazing areas for cattle and horses that are rich in annual vegetation (longitude 35°27’48’’E, latitude 33°04’56’’N, altitude 665 m above sea level) Degani [7]. The pools are filled with runoff water; water also seeps in from the settlements’ barns and coops. The pool is divided into a deeper part, about 2 m in depth, covering a total area of about 50 m2 (Figure 2) which holds water from around January to February Degani & Ahkked [2]; and the larger and shallower part, where the depth reaches about 80 cm at the center, the total area covers about 1000 m2 and holds water from January to June. In both parts of the pool, aquatic vegetation develops in the water: the common water-crowfoot (Ranunculus peltatus) and the common spike-rush (Eleocharis palustris).
Sample collection
The study was approved by the Israel Nature and National Park Protection Authority (permit 2020/42661). The pool area and its surroundings were explored from October to May Degani & Ahkked [2] (Figure 2). While the pool was dry, the entire area was examined once a week for newts in the terrestrial phase hiding under stones (during December 2020). When the pool was filled with water, aquatic adult newts (in January-February 2021) and larvae (in April 2021) were collected from it using a round hand net 40 cm in diameter with a mesh size of 0.1 cm. The net was immersed 40 cm into the water and 3–4 rotational movements of about 1 m from side to side were performed. All terrestrial and aquatic newts and aquatic larvae were released back into their respective habitats after being measured, photographed, and identified to the species level Degani & Ahkked [2]. A total of 20 tail samples (clipped tips of tails) were taken from 4 terrestrial females, 6 terrestrial males, 4 aquatic males, 1 aquatic female and 5 aquatic larvae and frozen in 1.5-ml tubes with RNA Later (Thermo Fisher Scientific) at −20˚C until further analysis.

RNA Extraction
Tissue samples were removed from RNA Later and homogenized using a Tissue Ruptor (Qiagen). Total RNA was extracted from each sample with TRI Reagent (Sigma) using the manufacturer’s protocol. The concentration and integrity of RNA were examined using a Thermo-Fisher Scientific Nano Drop 8000 Spectrophotometer and an Agilent 4150 Tape Station. 13 RNA samples had OD260/280 ≥ 1.8 and RNA integrity number (RIN) ≥ 7, and these were selected for further analysis.
Transcriptome Analysis
RNA-seq libraries were prepared at the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science using the in-house polyA-based RNA seq protocol (INCPM mRNA-Seq). Briefly, the polyA fraction (mRNA) was purified from 500 ng of total input RNA followed by fragmentation and generation of doublestranded cDNA. After cleanup with Agincourt AM Pure XP beads (Beckman Coulter), end repair, A base addition, adapter ligation and PCR amplification steps were performed. Libraries were quantified by Qubit (Thermo Fisher Scientific) and Tape Station (Agilent). Sequencing libraries were constructed with barcodes to allow multiplexing of 24 samples on an Illumina Nova Seq 6000 machine, using an SP (100 cycles) kit. 100bp single reads were sequenced on 2 lanes. The output was between 13-40 million reads per sample. Fast files for each sample were generated by bcl2fastq v2.20.0.422. Poly-A/T stretches and Illumina adapters were trimmed from the reads using cut adapt; resulting reads shorter than 30bp were discarded. Trimmed reads were used for assembly using Trinity (v2.13.2) Grabherr [10]. After the assembly, read representation was assessed with Bowtie2 (v2.3.4.3) Langmead [11] and completeness was assessed using BUSCO (v5.4.4) Manni [12]. Clustering of reads was performed using CD-hit (v4.8.1) Fu [13]. Trans decoder (v5.7.0) was used to predict coding regions from the long transcript of each gene. Eggnog-mapper was used to add functional annotations to the proteins’ sequences. The resulting annotation was merged with the DE analysis results (below).
Differential Expression Analysis
Differentially expressed genes were identified using DESeq2 Love [14] with the beta Prior, cooks Cutoff and independent Filtering parameters set to False. Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg Benjamini [15]. Pipeline was run using snake make Köster & Rahman [16]; Love [14]. The 1000 most variable genes from the DESeq2 analysis served for Principal Component Analysis (PCA) and Hierarchical Clustering (HCL) plots using default values. Volcano plots were prepared using the plot R package using DESeq2 output.
GO and pathway analysis
Functional enrichment analysis was performed using g_ Profiler (version e109_eg56_p17_1d3191d) with g:SCS multiple testing correction method, applying a significance threshold of 0.05 (Raudvere, Kolberg et al 2019).
Results
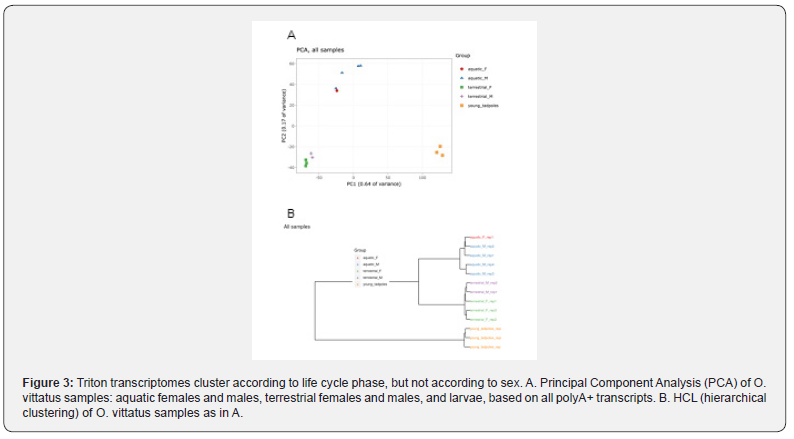
The analysis of the transcriptome variation in the tail tissues at the different phases of the newt life cycle shows clear and distinct differences between the gene expression of 3 groups: 1 – terrestrial newts (males and females), 2 - aquatic newts (males and females) and 3 – aquatic larvae before metamorphosis. The clustering of samples according to life cycle phase, but not according to sex, is apparent in PCA (Figure 3) and hierarchical tree analysis (Figure 3). De-novo transcriptome assembly from all samples resulted in a total of 143,575 assembled transcripts; 116,464 “genes” were represented. BUSCO completeness analysis searched a total of 255 groups, of which 249 (97.6%) were complete, 151 (59.2%) were also single-copy, 98 (38.4%) were duplicated, 6 (2.4%) were fragmented, and 0 were missing. BLAST hits were observed for 31,905 genes (min e-value was used to select the best hit). Of them, 15,299 (48%) were classified as bacterial and were filtered out. 12,117 hits belonged to eukaryotes. Of these, 3388 were similar to known avian genes, 2893 to mammals, 1670 to amphibians, 514 to bony fishes, and 239 to other eukaryotes.
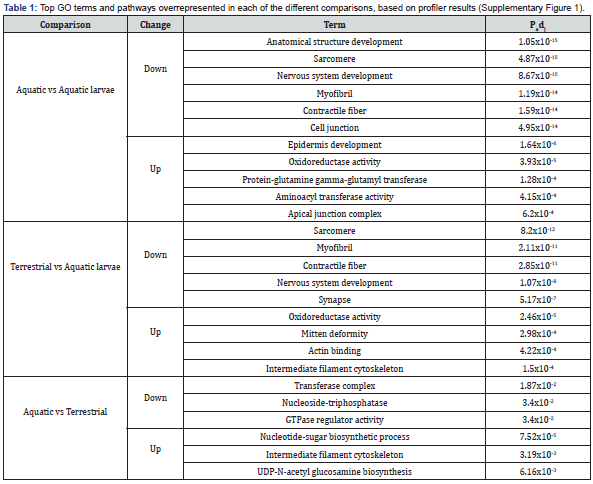
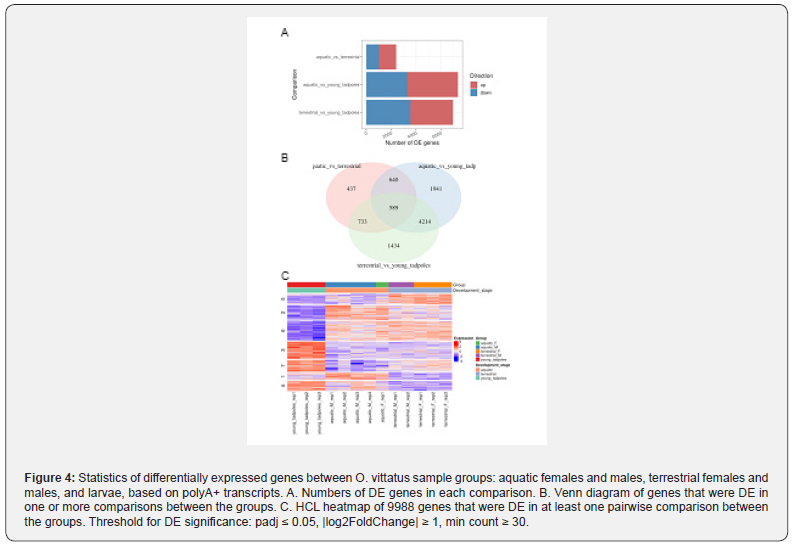
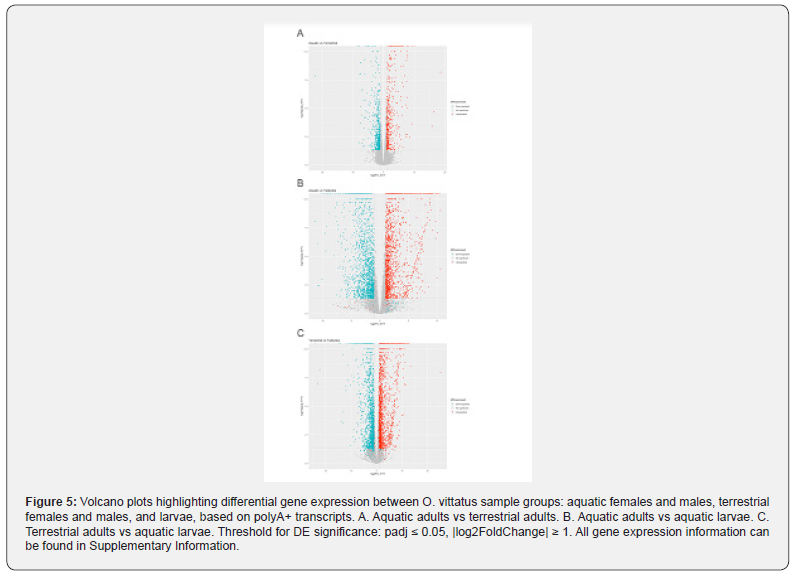
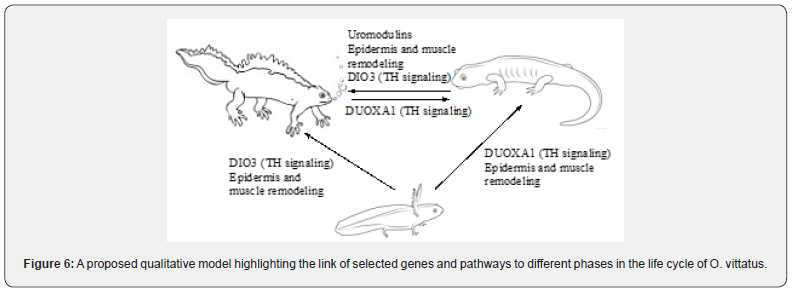
To assess differential expression (DE) of genes between the biological groups, reads from the individual samples were aligned to the assembly. The largest number of DE genes was found between aquatic larvae and adult newts (~7K-7.4K DE genes); comparing the aquatic and terrestrial adult groups showed ~2.4K DE genes (Figure 4). This was also apparent when looking at the changes in individual genes (volcano plots, (Figure 5). The most prominent DE genes in all comparisons (DE analysis output, Supplementary Information) were those coding for different variants of keratins and homologs of uromodulin. The latter were dramatically downregulated (by 4-5 orders of magnitude in expression) in terrestrial adults vs. aquatic larvae, and likewise dramatically upregulated in aquatic vs. terrestrial adults. We further screened DE genes for gene names and pathways previously reported to take part in amphibian metamorphosis. Homologs of several known genes involved in thyroid hormone signaling were identified in the newt transcriptome (Supplementary Information); among them, DUOXA1 (Dual Oxidase Maturation Factor 1) and PTH1R (Parathyroid Hormone 1 Receptor) were downregulated ~3- fold (padj=0.024) and ~2.6 fold (padj= 0.0012), respectively, while DIO3 (Iodothyronine deiodinase) was upregulated ~3- fold (padj= 0.0059) in aquatic vs. terrestrial adults. In agreement with a consistent role in the aquatic-terrestrial switch, DUOXA1 was also upregulated ~8.5-fold (padj= 6.7E-08) in terrestrial adults vs. aquatic larvae; while DIO3 was upregulated 6.6-fold (padj= 0.000002) in aquatic adults vs. aquatic larvae, indicating an association with metamorphosis. To gain insights into the functions of the DE genes, we performed g_profiler GO and pathway analysis. The top GO terms and pathways for the different comparisons are presented in Table 1 and Figure 6; further detail is provided in Supplementary Information. The samples examined in the different phases was from the newt’s tail. Thus, it is not surprising that the GO terms and pathways overrepresented in the different comparisons, are mainly related to structural anatomy and connective tissues, inter-cell connections, muscle and nerve cell generation, etc. Overrepresented developmental and metabolic pathways included “epidermis development”, “nervous system development”, “nucleotide-sugar biosynthesis”, “oxidoreductase activity”, potentially indicating the importance of these functions in metamorphosis and adaptation.
All gene expression information can be found in Supplementary Information.
Discussion
The life cycle, behavior and genetic variations among O. vittatus populations in northern Israel down to the central coastal plains and near the desert, were described previously; see review, Degani [9]. Our study found transcriptomic differences not only between aquatic larvae and adult newts, but also between the terrestrial and aquatic adult phases characteristic of the genus Ommatotriton Van Riemsdijk [17]; Van Riemsdijk [3]. Until recently, most “omics” studies of the genus Ommatotriton focused on differences between populations Van Riemsdijk [17]; Van Riemsdijk [3] and in phylogenetic aspects, unlike our study which for the first time examined the developmental changes in gene expression in an Ommatotriton species. We identified DE genes between the different life phases of the newts, namely aquatic larvae and adults in aquatic and terrestrial phases. The overall difference in gene expression between the aquatic larvae and adults is greater than the difference between the terrestrial and aquatic phases, which mirrors the great physiological changes in metamorphosis Degani [7]; Degani & Ahkked [2] and is in agreement with studies in other amphibians which undergo metamorphosis. Thus, in leopard frogs (Lithobates sphenocephalus) 42% of genes were differentially expressed between aquatic larvae and juveniles (Schott et al., 2022). Similar findings were reported in other newt species, e.g. the ribbed newt (Pleurodeles waltl) Matsunami [18].
We also identified DE genes between the aquatic and terrestrial phases in adult newts, which to the best of our knowledge has not been described in O. vittatus or other newts. Thus, uromodulin’s being the main protein secreted by the kidneys, the differential expression of uromodulin homologs between aquatic and terrestrial phases of the newt is in agreement with the importance of changes in kidney function between aquatic and terrestrial environments. It is well-known that the pattern of nitrogen excretion changes in the metamorphosis from aquatic larva to terrestrial adult, and also in the switch between terrestrial and aquatic adult phenotypes Nash & Frankhauser [19]. Thus, ~75 % of the nitrogen is excreted as ammonia and 25% as urea in the larval stage; terrestrial adults excrete ~87% of the nitrogen as urea; and aquatic adults partially reverse this change, doubling ammonia excretion to 26% Nash & Frankhauser [19]. Additionally, we report the association of elevated DUOXA1 (Dual Oxidase Maturation Factor 1), a positive modulator of thyroid hormone signaling (Szanto et al., 2019), with the terrestrial phenotype. Thyroid hormone signaling is a key trigger of amphibian metamorphosis Brown & Cai [20], and our findings suggest that it may also play a part in the aquatic-terrestrial switch in adult newts (Figure 6).
Our study was limited by the sample size, which was a consequence of the difficulty of capturing newts in the wild. Additionally, ethical considerations dictated that only tail samples be taken. Thus, the observed changes in gene expression may not be representative of those changes that are most functional in the developmental and physiological processes underlying the phenotypes [21,22]. For example, we would expect the kidneys to produce a clearer picture of the gene expression changes most relevant to the terrestrial-aquatic switch. However, the large number of DE genes identified, and the probable functional role of a subset of them based on prior knowledge, indicate that the use of tail section sampling, although only partially representative, is nevertheless warranted and informative. Future research, to include qRT-PCR validation of changes in the abundance of specific candidate transcripts and in specific tissues, could further elucidate the molecular mechanisms underlying newt development and adaptation. As climate change inevitably impacts the yearly wet-dry cycle of many habitats, the adaptability of the newt to aquatic and terrestrial lifestyles becomes paramount to its survival. Understanding the roles of specific genes and pathways in such adaptation, and assessing the variability in these traits between and within populations, can help predict population dynamics in vulnerable habitats and direct conservation efforts.
Conclusions
Both metamorphosis and environmental adaptation of banded newts involve extensive transcriptomic remodeling involving developmental, metabolic, and cellular pathways.
Understanding the roles of these pathways and individual genes is instrumental for studies aimed at conservation and preservation, especially in habitats affected by climate change. Furthermore, the phenotypic flexibility of the newt and the underlying regulation of gene expression can shed light on the evolution of terrestrial vertebrates.
References
- Degani G (2019) The Fire salamandra (Salamandra infraimmaculata) and the Banded newt (Triturus vittatus) along the southern border of their. Published by Scientific Research Publishing Inc.
- Degani G, Ahkked N (2021) Ecological and Biological Adaptations of Triturus vittatus vittatus (Urodela) to an Unstable Habitat. Int J Zoo Animal Bio 3: 1-8.
- Van Riemsdijk I, Arntzen JW, Bogaerts S, Franzen M, Litvinchuk SN, et al. (2017) The Near East as a cradle of biodiversity: A phylogeography of banded newts (genus Ommatotriton) reveals extensive inter- and intraspecific genetic differentiation. Mol Phylo Evol 114: 73-81.
- Geffen E, Gafny S, Gasith A (1987) Contribution to the knowledge of the biology of the banded newt, Triturus vittatus vittatus, in rainpools in Israel. Israel Journal of Zoology 34: 213-223.
- Pearlson O, Degani G (2008) The life history of Triturus v. vittatus (Urodela) in various habitats. Asiatic Herpetological Research 11: 91-95.
- Degani G (1982) Amphibian tadpole interaction in a winter pond. Hydrobiologia 96: 3-8.
- Degani G (2018) Genetic Variation in Xeric Habitats of Triturus vittatus (Urodela) Using Mitochondrial DNA of 12S and16S, and Nuclear Gene, Rhodopsin, on the Southern Border of its Distribution International Journal of Zoological Investigations 4: 31-40.
- Damen M, Vignoli L, Bologna MA (2006) The normal development and the chromosome No. 1 syndrome in Triturus carnifex carnifex (Caudata, Salamandridae). Italian Journal of Zoology 73(4): 325-333.
- Degani G (2017) Ecological, Biological, Behavioral and Genetic Adaptation to Xeric Habitats of Triturus Vittatus (Urodela) on the Southern Border of its Distribution. J Marine Sci Res Dev 7: 9910-2155.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat biotechnol 29(7): 644-652.
- Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie. Nat Methods 9(4): 357-359.
- Manni M, Berkeley MR, Seppey M, Simao FA, Zdobnov EM (2021) BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Molecular Biology and Evolution 38(10): 4647-4654.
- Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28(23): 3150-3152.
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for rna-seq data with deseq. Genome Biology 15: 1-21.
- Benjamini Y (2010) Discovering the False Discovery Rate. Journal of the Royal Statistical Society Series B: Statistical Methodology 72(4): 405-416.
- Köster J, Rahmann S (2012) Snake make-a scalable bioinformatics workflow engine. Bioinformatics 28: 2520-2522.
- Van Riemsdijk I, Arntzen JW, Babik W, Bogaerts S, Franzen M, et al. (2022) Next-generation phylogeography of the banded newts (Ommatotriton): A phylogenetic hypothesis for three ancient species with geographically restricted interspecific gene flow and deep intraspecific genetic structure Molecular Phylogenetics and Evolution 167: 107-361.
- Matsunami M, Suzuki M, Haramoto Y, Fukui A, Inoue T, et al. (2019) A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology. DNA Res 26: 217-229.
- Nash G, Fankhauser G (1959) Changes in the Pattern of Nitrogen Excretion during the Life Cycle of the Newt. Science 130(3377): 714-716.
- Brown D, Cai L (2007) Amphibian metamorphosis. Dev Biol 306(1): 20-33.
- Pearlson O, Degani G (2007) Triturus v. vittatus (Urodela) larvae at various breeding sites in Israel. Progress and Perspectives in Veterinary Medicine - Scientific Papers 50: 214-226.
- Schott RK, Bell RC, Loew ER, Thomas KN, Gower DJ (2022) Transcriptomic evidence for visual adaptation during the aquatic to terrestrial metamorphosis in leopard frogs. Biology 20: 138.






























