Evaluation of Candidates MiRNAs as a Diagnostic and Therapeutic Biomarker Between HPV Infection and Cervical Cancer Invasion via In Silico Approach
Hybert Keshishian1, Hamid Taghvaei Javanshir2, Maryam Arabi3 and Ahmad Bereimipour4*
1Department of microbiology, Islamic Azad University, North Tehran Branch, Tehran, Iran
2Cancer research centre, Tehran medical science. University of Tehran, Tehran, Iran
3Department of cell biology, Islamic Azad University, Tehran medical science, Tehran, Iran
4Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
Submission: November 20, 2021; Published: November 29, 2021
*Corresponding Address: Ahmad Bereimipour, Department of Stem Cells and Developmental Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
How to cite this article: Hybert Keshishian, Hamid Taghvaei Javanshir, Maryam Arabi and Ahmad Bereimipour. Evaluation of Candidates MiRNAs as a Diagnostic and Therapeutic Biomarker Between HPV Infection and Cervical Cancer Invasion via In Silico Approach. Canc Therapy & Oncol Int J. 2021; 20(2): 556033. DOI:10.19080/CTOIJ.2021.20.556033
Abstract
Background: Cervical cancer is one of the most common cancers. Women. One of the factors involved in the development of this cancer is the persistence of HPV infection. There are many treatments for cervical cancer, but the exact link between HPV and the development of cervical cancer remains unclear. Therefore, finding diagnostic biomarkers will be helpful in this regard.
Materials and Methods: This study was based on bioinformatics. First, we selected the appropriate microarray datasets from the GEO database. We categorized the high and low expression genes separately and used the Enrichr and Shiny GO databases to examine the signal pathways and gene ontology. We examined the association between proteins and evaluated candidate proteins with GEPIA in clinical samples using the STRING database. Finally, with the use of Targetscan, selected miRNAs.
Results: 204 genes with high expression and 171 genes with low expression were classified. High-expression genes were observed in the autoimmunity, antigen processing and presentation, pathways, and low-expression genes were observed in the cellular aging, and MAPK pathways. After delineating the protein linkage network, high-expression FOXA1, OSA1, IRF3, and STAT1, and low-expression BMP4, MEF2C, TBX2 and PTN were selected. Later, hsa-miR-203a-3p, hsa-miR-6881-3p, hsa-miR-3173-3p, hsa-miR-4796-5p were more closely related to these genes.
Conclusion: Finally, based on integrated and systematic bioinformatics analyzes, we nominated genes, proteins and important myrrh among cervical cancer-related HPV infection as biomarkers that need further investigation.
Keywords: MicroRNAs; Biomarker discovery; Human papilloma virus; Cervical Cancer; Invasive; In Silico Analysis
Abbreviations: MiRNAs: micro RNAs; HPV: Human Papilloma Virus; CC: Cervical Cancer
Introduction
Cervical cancer is one of the five common cancers in women [1]. Also, the five-year survival rate of this cancer is proportional to various physiological and environmental variables between 60 and 70%. Multiple variables are involved in the development of cervical cancer [2]. One of these causes is human papillomavirus infection. Infection with the virus, more common in the genital area, can trigger cervical cancer if the infection is severe or even after treatment [3]. Various treatments have been used to treat cervical cancer [4]. But to better manage the treatment of this cancer and to find the main actors in the development of cervical cancer due to human papillomavirus infection, the study of biomarkers is a very good option today. Over the past decade, bioinformatics knowledge has been able to determine and facilitate a comprehensive profile of cellular and molecular events involved in the pathogenesis of various disorders in the body by examining clinical data and differentiating gene expression [5,6]. Therefore, we used bioinformatics analysis in this study to find diagnostic and even therapeutic biomarkers for cervical cancer due to human papillomavirus infection, and finally nominated biomarkers for further studies.
Materials and Methods
Select the appropriate database and prepare the data
To design this study with royan institute, which has been done in bioinformatics analysis, we first used the GEO database and selected the appropriate microarray-datasets for this study. GSE63678 dataset was related to the cervical cancer and control group, divided into two groups and evaluated. This dataset consisted of 35 samples. GSE65166 dataset was related to HPV infection and control group, and these two groups were evaluated together. This dataset also included 9 samples. The platforms used in these datasets were GPL571 [HG-U133A_2] Affymetrix Human Genome U133A 2.0 Array and GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version], respectively. After defining the groups using the GEO2R tool, we isolated the differential expression profiles of the genes and saved them in an Excel file. Then we separated the gene clusters with up and down expression and prepared them for the next step. In this part, the p value˂0.05 was considered.
Investigation of signaling pathways and gene ontology
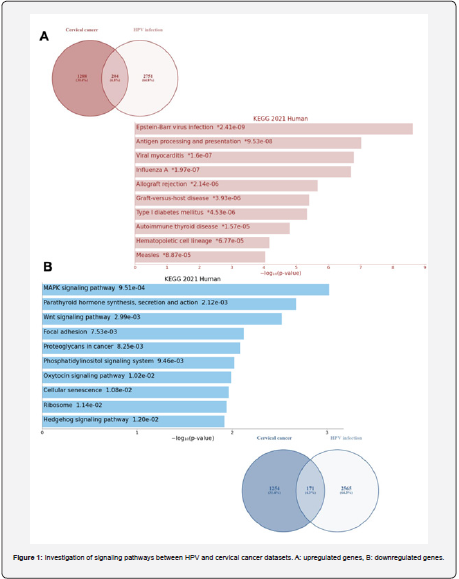
This section first used Venny version 2.1.0 to share genes with high and low expression of two datasets separately. We then uploaded the obtained common genes to the Enrichr database to examine the signaling pathways and gene ontology. Then used the KEGG and Reactions libraries to analyze the signaling pathways. The ontology section was then used to evaluate the molecular functions and biological processes of high and lowexpression genes. The Shiny GO database was then used to plot the communication network between the results. In this part, the p value˂0.05 was considered.
Evaluation of protein and gene networks
After evaluating the signaling pathways and gene ontology, the pathways that played a significant role in the development of cervical cancer dependent to HPV infection were selected. The relationship between their protein networks was assessed using the STRING database and the relationship between genes in the GeneMANIA database.
Evaluation of candidate genes and proteins in human data
After nominating important genes and proteins in the evaluated pathways, to confirm and obtain more information about the amount of expression, the algorithm for differentiating the expression of proteins in different stages of cervical cancer and survival based on time, the data in the GEPIA database uploaded and examined the parameters in patients with cervical cancer.
Select candidate microRNAs
After final confirmation and selection of genes and candidate proteins from the previous steps, we evaluated the microRNAs in the Enrichr database and the Targetscan library and selected high-significance microRNAs.
Results
Autoimmunity, antigen processing and presentation, cellular aging, and MAPK were more prominent between cervical cancer and HPV infection. After using the Venny tool to find the commonalities between the genes for the two datasets, 204 highexpression genes and 171 low-expression genes were obtained. Autoimmunity, antigen processing and presentation, Epsteinbar virus infection, allograft rejection, graft versus host disease, and hematopoietic cell lineage pathway were observed in highexpression genes, and cellular aging, cell cycle, MAPK, ribosome, Wnt signals, hedgehog signals, focal adhesion, and proteoglycan in cancer observed in low-expression genes (Figure 1).
Evaluation of gene ontology between cervical cancer and HPV infection
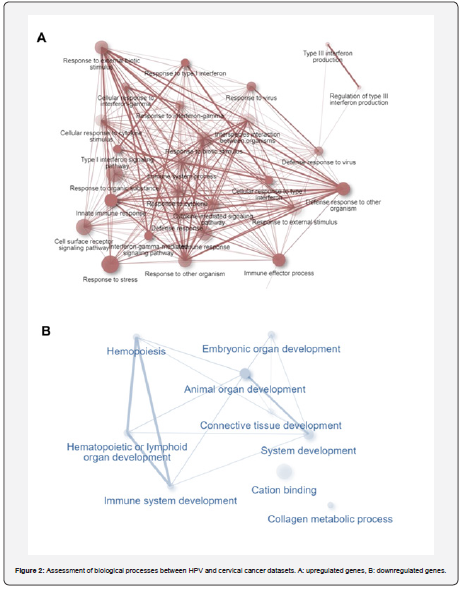
This section further evaluates the common genes from the previous step and examines their molecular functions and biological processes. Accordingly, type III interferon production, cellular response to type I interferon, defense response to virus, innate immune response, immune effector process. cell surface receptor signaling pathway, and detoxification of copper ion in biological processes were observed in genes with high expression. Also, for low-expression genes, blood vessel development, positive regulation of cellular component movement, tube development, regulation of cell population proliferation, positive regulation of RNA biosynthetic process, positive regulation of cellular biosynthetic process, regulation of RNA metabolic process and cellular developmental process were associated with biological processes (Figure 2).
Association between genes and proteins associated with the HPV infection and cervical cancer
The communication network between the proteins was plotted with high expression. The network consists of 83 nodes and 156 edges, with proteins that interact better with other proteins at the center of the network. The low expression proteins network consists of 61 nodes and 98 edges High-expression FOXA1, OSA1, IRF3 and STAT1 and low-expression BMP4, MEF2C, TBX2 and PTN proteins were selected more significantly (Figures 3A & 3B).

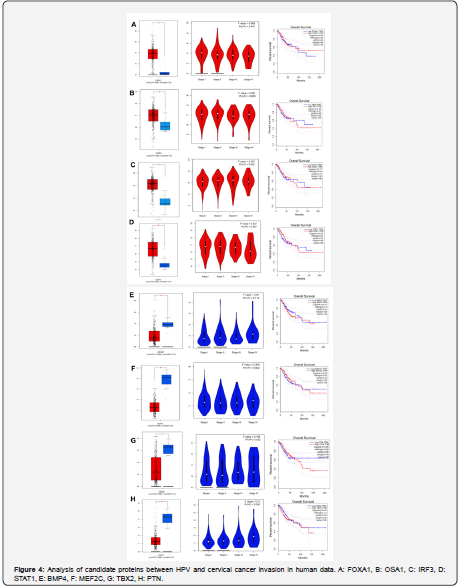
Evaluation of proteins in clinical data
In this part of the study, FOXA1, OSA1, IRF3, STAT1, BMP4, MEF2C, TBX2 and PTN proteins in the GEPIA database were evaluated in cervical cancer samples compared to controls. Accordingly, like bioinformatics data, thease proteins in the cervical cancer sample showed a significant increase and decrease in expression compared to the control sample. In the plot diagram, higher data density is directly related to increased gene expression. In the survival chart, on average, FOXA1, OSA1, IRF3, STAT1, BMP4, MEF2C, TBX2 and PTN proteins have reduced the survival of patients by about 50% over time, which is a significant rate (Figures 4 & 5).
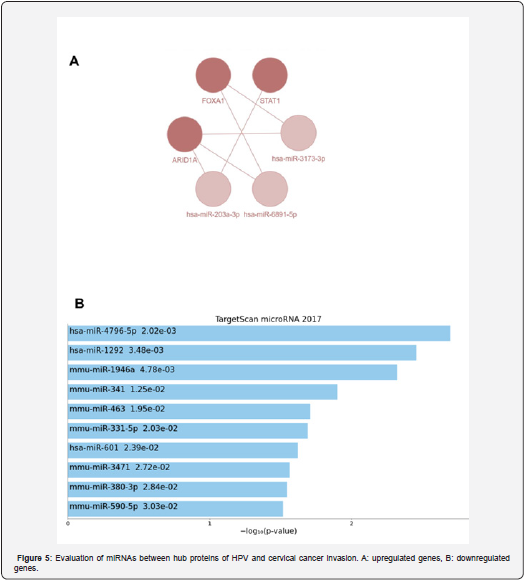
Candidate of suitable microRNAs related to the HPV infection and cervical cancer
In this section, we evaluated the candidate genes and proteins in terms of upstream regulatory elements. A communication network between microRNAs is also drawn. hsa-miR-203a-3p, hsa-miR-6881-3p, and hsa-miR-3173-3p regulated the highexpression genes and the hsa-miR-4796-5p, hsa-miR-1292, hsamiR- 1946a, hsa-miR-341 and hsa-miR-463 regulated the lowexpression genes (Figure 5).
Discussion
Cervical cancer is common cancer in women, and there are various challenges to developing this cancer. Of course, HPV infection is a particular variable that these viruses can cause cancer in the human body [7]. HPV16 and HPV18 more than other strains. HPV can cause cervical cancer. New methods to find the exact cause of the onset and development of cancers have become more prevalent in recent years [8]. Gene expression profile in cervical cancer and its association with HPV infection and finding diagnostic or therapeutic biomarkers can identify the key molecules in causing cervical cancer and increasing HPV infection to provide better treatment strategies [9].
Studies had shown that FOXA1 plays a vital role in inducing cell division and invasion of cervical cancer cells, controlled and regulated by miR 1179 [10]. Another study showed that FOXA1 is highly expressed in patients with cervical cancer. After inhibition of FOXA1 activity, invasion and division of cancer cells were significantly reduced. Cellular connections and EMT were also reduced [11]. The study by Karpathiou et al. Showed that FOXA1 expression was significantly increased in cervical and endocervical cancer patients compared to healthy individuals with HPV infection [12]. FOXA1 is also active in other cancers, including head and neck [12] and lung cancers [13].
Further studies based on bioinformatics and gene expression profile analysis in cervical cancer patients have shown that OAS1 plays a key role in the progression, cell division, and invasion of cervical cancer cells [14]. The study by Shah et al. With bioinformatics analysis showed that IRF3 plays an essential role in the response and resistance to drugs against HPV [15]. The study by Wang et al. Also showed that various mutations in the IRF3 gene disrupted DNA repair and DNA replication pathways in patients with HPV-associated cervical cancer [16]. An intriguing study by Cai et al. Showed that IFI16 also activated the STING / TBK1 / NFKB / IRF3 signal pathway when there was an increase in expression, which played a significant role in innate immunity as well as the progression of cervical cancer [17].
The study by Suk et al. Showed that IFN gamma could control the STAT1 / IRF1 signal pathway, which is involved in cell division and cervical cancer invasion [18]. The study by Wu et al. Also showed that STAT1 played an important role in cervical cancer and introduced it as a marker [19]. The study by Raspaglio et al. Showed that STAT1, controlled by PARP1, could be sensitive to radiotherapy in cervical cancer cells [20]. Zhang et al. showed that STAT1 controls p53, and when the STAT1 upstream gene, FRA1, is activated, it inhibits cell division and the Warburg effect in cervical cancer [21]. Buttareli et al. [22] Also showed that the ANXA2 / NDRG1 / STAT1 signal axis plays an important role in resistance to chemotherapy in cervical cancer.
A recent study by Wang et al. Showed that miR 592 / RSPO1 by controlling MEF2C could reduce cell division and invade cervical cancer [23]. Another study showed that MEF2C increased expression in people with endometriosis compared to healthy people and was effective in the progression and exacerbation of the disease [24]. An intriguing study by Zhang et al. Showed that the MUC4 / ERbB2 / MEF2C / P38 signal pathway axis in pancreatic cancer is disrupted by decreased expression of MMP10 and is involved in the invasion and metastasis of this cancer [25].
The study by Liu et al. Showed that PSCA, PIWIL1, and TBX2 genes have significantly increased expression in patients with cervical cancer, which has a significant role in cell division, invasion, and progression of this cancer, especially to lymph nodes [26]. The study by Schneider et al. Showed that TBX2 and TBX3 are directly related to L2 from HPV and play an important role in regulating the viral cycle in the host cell. It can also control and regulate viral infection in the host body in the long run. This factor can play a key role in exacerbating cervical cancer [27]. One study found that when curcumin was added to cervical cancer cells, PTN expression was significantly reduced, reducing cell division and invasion of cervical cancer cells [28].
Conclusion
Finally, in the present study, we examined the miRNAs associated with these genes, and finally, the hsa-miR-203a-3p, hsa-miR-6881-3p, hsa-miR-3173-3p, hsa-miR-4796-5p, hsamiR- 1292, hsa-miR-1946a, hsa-miR-341 and hsa-miR-463 were selected. Finally, a more in-depth study of genes and miRNAs involved in HPV-associated cervical cancer could provide specialists with better treatment options.
Conflict of Interest
The authors declared that they have no conflict of interest.
Acknowledgement
We thank all colleagues in the core facilities for their support.
References
- Lei J, Ploner A, Elfstrom KM, Wang J, Roth A, et al. (2020) HPV vaccination and the risk of invasive cervical cancer. N Engl J Med 383(14): 1340–1348.
- Kashyap N, Krishnan N, Kaur S, Ghai S (2019) Risk factors of cervical cancer: a case-control study. Asia-Pacific J Oncol Nurs 6(3): 308.
- Chrysostomou AC, Stylianou DC, Constantinidou A, Kostrikis LG (2018) Cervical cancer screening programs in Europe: the transition towards HPV vaccination and population-based HPV testing. Viruses. 10(12): 729.
- Beharee N, Shi Z, Wu D, Wang J (2019) Diagnosis and treatment of cervical cancer in pregnant women. Cancer Med 8(12): 5425–5430.
- Chen L, Heikkinen L, Wang C, Yang Y, Sun H, et al. (2019) Trends in the development of miRNA bioinformatics tools. Brief Bioinform 20(5): 1836–1852.
- Chen L, Wang C, Sun H, Wang J, Liang Y, et al. (2021) The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 22(2): 1706–1728.
- Davies-Oliveira JC, Smith MA, Grover S, Canfell K, et al. (2021) Eliminating cervical cancer: progress and challenges for high-income countries. Clin Oncol 33(9): 550–559.
- Clifford GM, Tenet V, Georges D, Alemany L, Pavon MA, et al. (2019) Human papillomavirus 16 sub-lineage dispersal and cervical cancer risk worldwide: Whole viral genome sequences from 7116 HPV16-positive women. Papillomavirus Res 7: 67–74.
- Wu X, Peng L, Zhang Y, Chen S, Lei Q, et al. (2019) Identification of key genes and pathways in cervical cancer by bioinformatics analysis. Int J Med Sci 16(6): 800.
- Yang S, Jiang Y, Ren X, Feng D, Zhang L, et al. (2020) FOXA1-induced circOSBPL10 potentiates cervical cancer cell proliferation and migration through miR-1179/UBE2Q1 axis. Cancer Cell Int 20(1): 1–12.
- Gu F, Li ZH, Wang CQ, Yuan QF, Yan ZM (2018) Effects of forkhead Box protein A1 on cell proliferation regulating and EMT of cervical carcinoma. Eur Rev Med Pharmacol Sci 22: 7189–7196.
- Karpathiou G, Da Cruz V, Casteillo F, Mobarki M, Dumollard JM, et al. (2017) FOXA1 in HPV associated carcinomas: Its expression in carcinomas of the head and neck and of the uterine cervix. Exp Mol Pathol. 102(2): 230–236.
- Ye J-J, Cheng Y-L, Deng J-J, Tao W-P, Wu L (2019) LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma by targeting miR-302c-5p/FOXA1 axis. Gene 685: 76–84.
- Zhang X, Yang P, Luo X, Su C, Chen Y, et al. (2019) High olive oil diets enhance cervical tumour growth in mice: transcriptome analysis for potential candidate genes and pathways. Lipids Health Dis 18(1): 1–13.
- Shah M, Anwar MA, Park S, Jafri SS, Choi S (2015) In silico mechanistic analysis of IRF3 inactivation and high-risk HPV E6 species-dependent drug response. Sci Rep 5(1): 1–14.
- Wang SS, Bratti MC, Rodriguez AC, Herrero R, Burk RD, et al. (2009) Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J Infect Dis 199(1): 20–30.
- Cai H, Yan L, Liu N, Xu M, Cai H (2020) IFI16 promotes cervical cancer progression by upregulating PD-L1 in immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed & Pharmacother 123: 109790.
- Suk K, Chang I, Kim Y-H, Kim S, Kim JY, et al. (2001) Interferon $γ$ (IFN$γ$) and tumor necrosis factor $α$ synergism in ME-180 cervical cancer cell apoptosis and necrosis: IFN$γ$ inhibits cytoprotective NF-$κ$B through STAT1/IRF-1 pathways. J Biol Chem 276(16): 13153–13159.
- Wu S, Wu Y, Lu Y, Yue Y, Cui C, et al. (2020) STAT1 expression and HPV16 viral load predict cervical lesion progression. Oncol Lett 20(4): 28.
- Raspaglio G, Buttarelli M, Filippetti F, Battaglia A, Buzzonetti A, et al. (2021) Stat1 confers sensitivity to radiation in cervical cancer cells by controlling Parp1 levels: a new perspective for Parp1 inhibition. Cell Death & Dis 12(10): 1–13.
- Zhang M, Liang L, He J, He Z, Yue C, et al. (2020) Fra-1 Inhibits Cell Growth and the Warburg Effect in Cervical Cancer Cells via STAT1 Regulation of the p53 Signaling Pathway. Front Cell Dev Biol 8: 1022.
- Buttarelli M, Babini G, Raspaglio G, Filippetti F, Battaglia A, et al. (2019) A combined ANXA2-NDRG1-STAT1 gene signature predicts response to chemoradiotherapy in cervical cancer. J Exp & Clin Cancer Res 38(1): 1–17.
- Wang X, Zhang C, Gong M, Jiang C (2021) A Novel Identified Long Non-coding RNA, lncRNA MEF2C-AS1, Inhibits Cervical Cancer via Regulation of miR-592/RSPO1. Front Mol Biosci 8: 544.
- Gao C, Zhou C, Zhuang J, Liu L, Liu C, et al. (2018) MicroRNA expression in cervical cancer: novel diagnostic and prognostic biomarkers. J Cell Biochem 119(8): 7080–7090.
- Zhang J-J, Zhu Y, Xie K-L, Peng Y-P, Tao J-Q, et al. (2014) Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol Cancer 13(1): 1–17.
- Liu W-K, Jiang X-Y, Zhang Z-X (2010) Expression of PSCA, PIWIL1 and TBX2 and its correlation with HPV16 infection in formalin-fixed, paraffin-embedded cervical squamous cell carcinoma specimens. Arch Virol. 155(5): 657–663.
- Schneider MA, Scheffer KD, Bund T, Boukhallouk F, Lambert C, et al. (2013) The transcription factors TBX2 and TBX3 interact with human papillomavirus 16 (HPV16) L2 and repress the long control region of HPVs. J Virol 87(8): 4461–4474.
- Ting-ting Fan, Aixue Liu, Ruinian Zheng, Lingyan Zhang, Li Huang, et al. (2008) Effect of curcumin on the protein expression of pleiotrophin in cervical cancer caski cells [J]. J Chongqing Med Univ 7.






























