Preprocessing of Artifacts Removal from Skull
Sudipta Roy1 and Samir Kumar Bandyopadhyay2
1Department of Computer Science and Engineering, Institute of Computer Technology (UVPCE), Ganpat University, India
2Advisor to Chancellor, JIS University, India
Submission: January 17, 2018; Published: March 16, 2018
*Corresponding author: Samir Kumar Bandyopadhyay, dvisor to Chancellor, JIS University, India, Email: 1954samir@gmail.com
How to cite this article: Sudipta R, Samir K B. Preprocessing of Artifacts Removal from Skull. Curr Trends Biomedical Eng & Biosci. 2018; 12(5): 555850. DOI: 10.19080/CTBEB.2018.12.555850
Abstract
Diagnosis quality of brain MR of brain images hampered due to the presence of artifacts. Small abnormalities detection hampered due to the presence of skull region of the brain. Sometimes artifacts and skull have been treated as an abnormality in the automated system, and it hampers the intelligence system. Thus a computerized method requires pre-processed image as artifacts and skull removal. Preprocessing makes the image segmentation more accurate. In this paper pre-processing method for improvement of brain abnormality detection and diagnosis has been described.
Keywords: Skull; CSF; Binarization; Artifacts
Introduction
Automatic segmentation of brain tissues from MRI is a challenging process due to the variation in brain shapes and similarity of intensity values in the brain and non-brain tissues. Artifacts and undesired tissues (skull) as non-brain region affect the quality of processing and may lead to automatic diagnostic confusion. It is done for the purpose of clearing away non-brain backgrounds and reducing unwanted information from the MR images.
The majority of skull stripping treats the brain as a single connected region separated from non-brain tissues by a rim of CSF. In reality, even with high-resolution, T1 weighted MR images, thin connections between the brain and other cranial structures exist in the form of dura and connective tissue lining venous sinuses.
The proposed preprocessing is very simple technique using the combination of binarization, wavelet decomposition, and computational geometric as major intermediate steps. It tested with a large number of MR images and produced good results. In preprocessing, method artifacts removal is considered as mandatory, and skull removal is optional. Artifacts removal is required for every post-processing technique, and skull removal is required for some abnormality detection like edema, tumor, stroke lesions and hemorrhage lesions, but sometimes for a particular type of hemorrhage, we need the distance of abnormal region from the skull.
Literature Review
Brain tumor, stroke, hemorrhage and multiple sclerosis (MS) disease are the life threatening diseases in both male and female. A brain tumor is the most common and widespread disease among these brain diseases. The worldwide cancer incidence of brain tumor is 3.4 per 100,000 people (men: 3.9 per 100,000, women: 3.0 per 100,000. A total of 256,213 affected worldwide (139,608 men and 116,605 women). The trend of new cases is rising and 189,582 sufferers worldwide. Every day about 700 people is diagnosed with a brain tumor [1]. 15 million people are affected by stroke and hemorrhage; of this 5 million die and another 5 million (2002 estimates) are permanently disabled. Today over 2,500,000 people around the world have MS [2].
Some automated and semi-automated skull stripping algorithms are available in the literature. Skull removal using graph cuts [8] relies on graph theoretic image segmentation techniques to position cuts that serve to isolate and remove skull. Region growing based [7] method for coronal T1- weighted images plans to automatically detect two seed regions of the brain and non-brain by using the mask generated by mathematical morphological operations. Then the seed regions were expanded using 2D region growing algorithm. Another fully automatic brain extraction method [8] using diffusion, run length encoding, and region labeling was developed for skull removal in T2 weighted axial MR brain images.
Clustering and 2D Region Growing method [9] for detecting the brain boundaries inside the skull was used to join the clusters and also remove the skull area. Authors are tested on four slices only thus method needed to improvement to process all the slices in the dataset. Robex method [10] for skull stripping by using a shape model trained on healthy brains to be relatively insensitive to lesions inside the brain. Their results showed this method was better than Brain Surface Extractor (BSE), hybrid watershed algorithm (HWA) and brain extraction tool (BET). They used T1 weight images taken from glioblastoma patients. A method based [11] on histogram analysis and compared the segmentation accuracy between their proposed method and two widely used techniques, namely BSE and BET. Based on this factor, they reported that their proposed method outperforms these methods.
A deep convolutional learning [12] architecture is used to skull removal but not limited to non-enhanced T1 images. When trained appropriately, it handles an arbitrary number of modalities including contrast-enhanced scans. Its applicability to MRI data, comprising four channels: non-enhanced and contrast-enhanced T1w, T2w and FLAIR contrasts, is demonstrated on a challenging clinical data set containing brain tumors. A mathematical morphology method was implemented [13] for the preprocessing of MR brain images for the improved segmentation of brain tumor based on mathematical morphology operations. The first part of that paper [13] was an efficient method for the skull stripping of brain MR images based on mathematical morphology. In that paper, brain part was identified by the largest connected component in the image after binarization. The largest connected component is then dilated with a 3x3 square structuring element so as to preserve minute brain information in the output image. The holes in the resultant image were filled to make the brain a complete connected component. The resulting pixels are superimposed with the input image for getting the skull removed image. But the problem arises when skull and brain is connected then that method fails to identify the difference between brain and skull. Most of these methods apply to specific a type of MR brain images and do not extract the brain completely in all the slices.
Moreover, none of these presented methods give good performance when evaluated for large-scale data set. It is due to the complexity of the human brain, varying image contrast properties, image artifacts such as under-sampling, noise factor, variations in the image orientations and types.
Data Sets
The proposed techniques implemented based on brain MRI images consisting of normal and abnormal from a real human brain MRI dataset. The dataset used consists of axial, T2, T1, and PD MR Brain Images. These images were collected from the Harvard Medical School [5] website used for normal and abnormal brain images: (a) tumors, (b) strokes, (c) hemorrhage, and (d) MS. Standard simulations from brain web database [6] include parameter setting fixed to 3 modalities (T1, T2, and PD), five slice thicknesses (1, 3, 5, 7 or 9mm), and defining a volume (x, y, z). Tissue classes include GM, WM, and CSF but also muscle, fat or skin. The brain model used to generate the simulations can also be employed as ground truth. Another public domain dataset is used to conducting similar research. Easy Analytic Software, Inc (EASI) MRI database for different brain abnormality MR images [7]. All mentioned dataset are used to evaluate the performance. MRI is the most frequently used neuroimaging technique for the evaluation and follow-up a review of patients with brain abnormalities for many reasons [34]. It does not use ionizing radiation like computed tomography (CT), single photon emission computed tomography (SPECT), and positron emission tomography (PET) studies.

Proposed methodology
Artifacts removal is an essential task for normal and abnormality brain tissues identification. Skull elimination is another important step for abnormality segmentation. In the case of particular type of hemorrhage (e.g., chronic subdural hematoma) lesions segmentation skull information is required due to the distance measurement from the skull. Thus in preprocessing method skull removal is not mandatory for some few cases, it depends on applications. Figure 1 shows a flowchart of preprocessing of brain normal and abnormal tissue detection.
Artifacts and skull removal both need to perform following intermediate steps after performing above steps:
i. Complement the binarized image that helps for wavelet decomposition because we want to disconnect main brain region from skull where both are connected.
ii. Two-dimensional wavelet decompositions are done using 'dbl' wavelet up to level two. The connection between brain and skull region are removed when we recompose the image.
iii. Re-composition of the image is done using the approximate coefficient of the previous step. Then resize the image of the previous step to the original size and recomplement of the image.
iv. Select a largest connected area of from connected components and remove other components. This largest component is the brain without skull region in MRI.
After performing the artifacts or artifacts and skull removal steps, we use some common procedure. The binary image contains white pixels are losses some information in the border as well as inside the bounded area. To make it perfect as possible we perform quickhull [14-15] on white pixels. The quickhull algorithm uses less space than most of the randomized incremental algorithms and executes faster for inputs with nonextreme points. Computation time is less, and also, quickhull uses merged facets to guarantee that the output is clearly convex. Then that convex binarized image is multiplied with the original image to produce the final results.
The time complexity regarding big-O for proposed preprocessing method has been described. Assuming input image has m number of row and n number of column and if some row=number of column= n then to compute the binarized image O(n2)time required. Selection of maximum area require O(n) time, and convex hull computation takes O(n*n logn) time, and multiplication of each image pixel require O(n2) time. Thus the total time complexity: T(n) = O(n2) + O(n2) + O(n*n logn) + O(n) « O(n2 logn).
Results and Discussions
Proposed method gives suitable results for different MRI of brain images in artifacts removal as well as artifacts and skull removal. The results below have been tested visually and metrically. The procedure of proposed methods has been described above, and figures below show each functional step of the proposed methods. Figure 2 below shows the intermediary results of different intermediate steps of artifacts and skull removal methodology. Brain MRI image (a) is taken as input and converted it to a grayscale image for next step binarization. (b) Is the binarized output using the standard deviation based threshold intensity for the input image (a), here maximum of brain region along with artifacts are very clearly visible in the binary image. This visibility of maximum brain tissues and artifacts are very useful for next steps. It is clear that the ratio of brain to artifact gives very high (i.e. above 30). The ratio between the skull and internal brain tissue will not exceed 30 in any case. So, depending on the ratio between highest to second highest component brain and skull has been selected. (c)
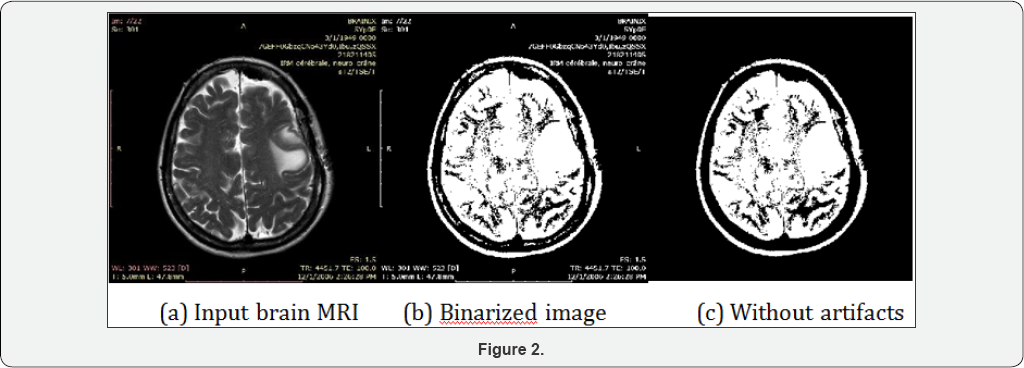
Artifacts removal technique can remove the artifacts if any artifacts present in the brain MRI. Proposed method is tested on a large dataset and produce excellent results except for connected artifact with the original brain portion image. Proposed technique is very helpful in the sense of brain tissues detection. Artifacts and skull removal technique is also to remove skull and artifacts for different images. The correct elimination of skull and artifacts will reduce false detection in abnormal tissues detection. Measure the performance by visually may be biased. Thus some performance evaluation metrics are used to evaluate the error and accuracy with respect to the reference image. The accuracy is used to evaluate the performance of the proposed methods are the Relative area Error (RAE or RE), Kappa Index (KI), Jacard Index (JI), Correct Detection Ratio (CDR) and False Detection Ratio (FDR). A critical problem faced in performance evaluation of artifacts and skull removal method is the lack of a gold standard. Here we use ground truth suggested by a radiologist for the comparison with the automated method and measures their performance with the help of RE, KI, JI, CDR, FDR. Let AS, and MS denote the area of the automatically segmented (AS) and manually segmented (MS) regions of the MR brain images. The Relative Error (RE) for the segmented region can be calculated as the difference between 'AS' and 'MS' by 'MS.'
Conclusion
In this paper, a fully automatic method for artifacts and skull removal of brain MR images using computational geometry, wavelet decomposition, and thresholding as intermediate steps has been described. Thresholding using standard deviation method is the key intermediate step to correct preprocessing of MR images. The proposed preprocessing method has high accuracy and low error rate for different MR images. This pre-processing is used to reduce the false detection. Thus it increases the diagnosis quality of disease from MRI of the brain by a computer system. Thus reprocessing with low error and high accuracy does not hamper the post-processing of intelligence system. The proposed method is very useful and important preprocessing for correct automated detection of brain abnormalities. This preprocessing method is used in abnormalities detection, segmentation and classification and tissues identification in this research.
References
- http://globocan.iarc.fr/Default.aspx
- The Multiple Sclerosis International Federation (MSIF).
- Ei Sayed A, Tamer H, Abdel B (2010) Hybrid intelligent techniques for MRI brain images classification. Digital Signal Processing, Elsevier 20(2): 433-441.
- Ricci PE, Dungan DH (2001) Imaging of low- and intermediate-grade gliomas. Seminars in Radiation Oncoloy 11(2): 103-112.
- http://www.med.harvad.edu/AANLIB/home.html
- http://brainweb.bic.mni.mcgill.ca/brainweb/
- http://www.easidemographics.com/cgi-bin/dbmri.asp
- Park JG, Lee C (2009) Skull Stripping Based on Region Growing for Magnetic Resonance Images. Neuroimage 47(4): 1394- 1407.
- Somasundaram K, Kalaiselvi T (2010) Fully Automatic Brain Extraction Algorithm for Axial T2- Weighted Magnetic Resonance Images. Computers in Biology and Medicine 40(10): 811-822.
- Somasundaram K, Siva Shankar R (2010) Kull Stripping of MRI Using Clustering and 2D Region Growing Method. Image Processing NCIMP.
- Speier W, Iglesias JE, El-Kara L, Tu Z, Arnold C (2011) Robust Skull Stripping of Clinical Glioblastoma Multiforme Data. MICCAI 2011, Part III, Springer LNCS 14(Pt 3): 659-666.
- Balan AG, Traina AJ, Ribeiro MX, Marques PM, Traina C (2012) Smart Histogram Analysis Applied To The Skull-Stripping Problem In T1- Weighted MRI. Computers in Biology and Medicine 42(5): 509-522.
- Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K (2016) Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. Neuroimage 129: 460-469.
- Benson CC, Lajish V.L, Kumar R (2016) A Novel Skull Stripping and Enhancement Algorithm for the Improved Brain Tumor Segmentation using Mathematical Morphology. IJ Image Graphics and Signal Processing 7: 59-66.
- Bradford C, Dobkin P, Hannu H (1996) The Quickhull Algorithm for Convex Hulls. ACM Transactions on Mathematical Software 22(4): 469-483.






























