Pseudomonas- A potential Source for Drug Lead Discovery
Kirti Hira1 , AmeerBasha Shaik2, Santosh Kumar1 and Sajeli Begum Ahil1*
1Department of Pharmacy, Hyderabad Campus, India
2Maize Research Centre, ARI, India
Submission: July 03, 2017; Published: July 21, 2017
*Corresponding author: Sajeli Begum Ahil, Department of Pharmacy, BITS-Pilani, Hyderabad Campus, Jawahar Nagar, Shameerpet, Hyderabad-500078, Telangana State, India, Tel: +914066303578; Email: sajeli@hyderabad.bits-pilani.ac.in
How to cite this article: Kirti H, Ameer B S, Santosh K, Sajeli B A. Pseudomonas- A potential Source for Drug Lead Discovery. Curr Trends Biomedical Eng & Biosci. 2017; 5(5): 555674. DOI: 10.19080/CTBEB.2017.05.555674
Abstract
Microorganisms have always been considered as important resource in drug discovery. As there is always a need for medicines with higher efficacy and lesser side effects, pharmaceutical research based on natural sources still continues by playing a vital role in discovery and development of drugs. Among microorganisms, bacteria have been identified as a potential spring for identifying new chemical entities. Pseudomonas is a diverse genus of Gammaproteobacteria which exhibits remarkable genomic diversity and is well recognized for the production of a variety of secondary metabolites. Pseudomonads have been shown to produce siderophores, pyoverdins, lipopeptides, numerous non- ribosomally derived peptides, polyketides and small molecular weight antibiotics or phytotoxins which demonstrate remarkable biological activities. The pharmacological activities exhibited by metabolites of different Pseudomonas species are elaborated in this review to explore the scope of probing pseudomonas species for drug lead molecules.
Keywords: Pseudomonas; Secondary metabolites; Siderophores; Pyoverdins; Lipopeptides; Polyketides; Cyclic dipeptides; Antimicrobial; Antiinflammatory
Introduction
Since the very existence of human civilization natural sources have been considered as the basis for the treatment of several diseases. Basis of development of modern medicine remain rooted in traditional medicine and therapies which were based on natural products [1,2]. Discovery of new antibiotics such as penicillin from microorganisms has led to the expansion of pharmaceutical research based on microbial sources [3]. Among the microbial sources, bacteria with a small group of taxa have been recognized as potential source for the development of new drugs. Discovery of novel secondary metabolites from marine actinomycetes also opened a new dimension for product research [4].
Pseudomonas
Pseudomonas is a diverse genus of Gamma proteobacteria with more than 60 species, ubiquitously present in environment such as soil, water, plant surface and animals. Pseudomonas species exhibits remarkable ecological and metabolic diversity with complete genomes of at least 22 strains. They are well known for their enormous metabolic capability to produce remarkable array of structurally diverse bioactive secondary metabolites [5] (Figure 1). Pseudomonads are capable of producing siderophores, pyoverdins [6-8], lipopeptides [9], numerous non-ribosomally derived peptides [10,11], polyketides and small molecular weight antibiotics or phytotoxins [7,12,13].
Secondary metabolites of pseudomonas species and their biological activity
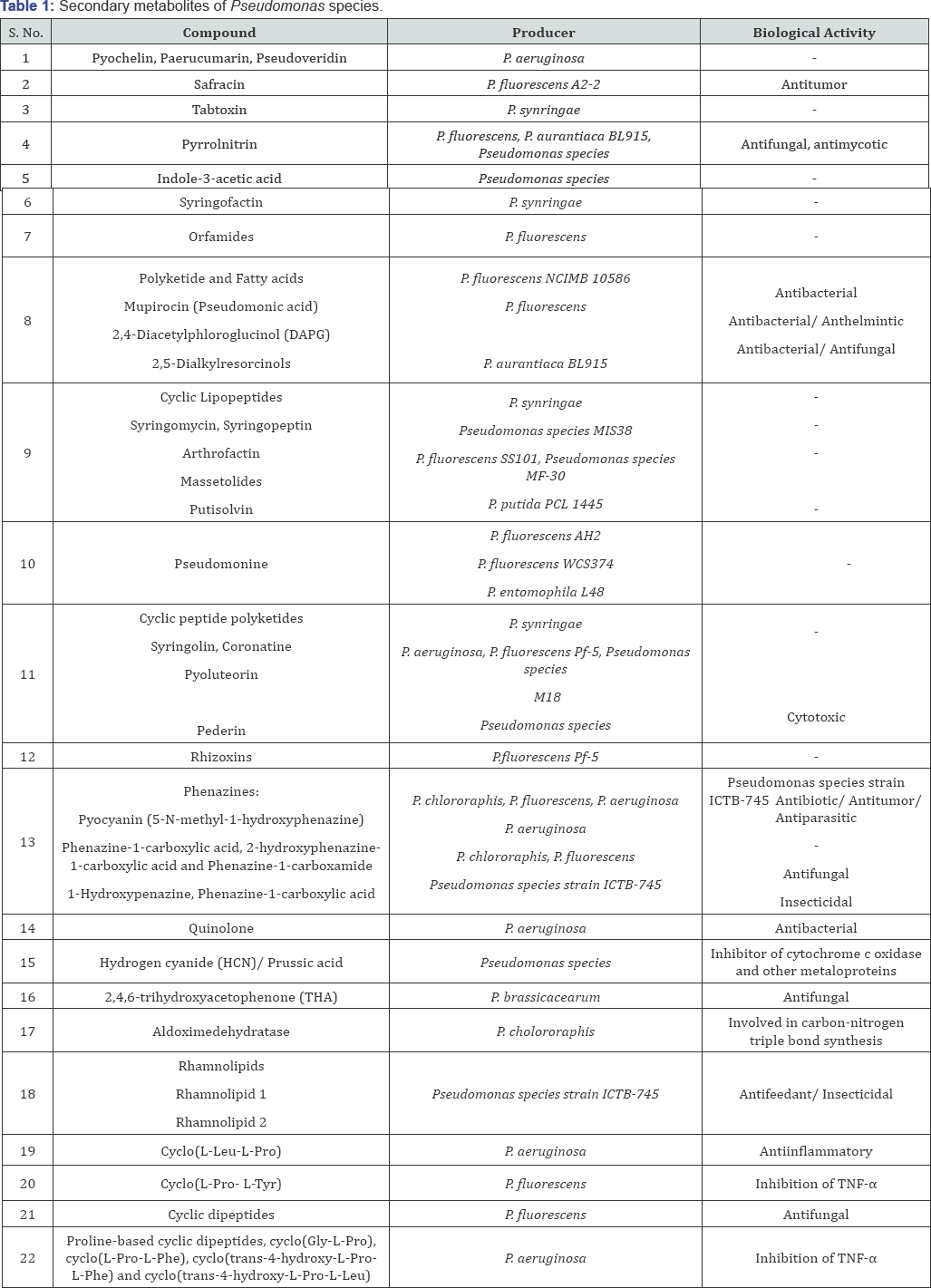
Pseudomonads are known to secrete numerous proteins and secondary metabolites that exhibit diverse biological activity [5,10,11,14-17]. Table 1 details the pharmacological effects of specific compounds produced by different species of Pseudomonas. Recent research is focused towards developing cyclic dipeptides produced by Pseudomonas as anti inflammatory molecules.
Acknowledgement
Authors acknowledge Department of Science and Technology, Govt. of India (EMR/2016/002460). One of the authors (KH) gratefully acknowledges Council for Scientific Industrial Research, India for grant of Junior Research Fellowship (09/1026(0019)/2017 EMR-I).
References
- Patwardhan B, Vaidya AD, Chorghade M (2004) Ayurveda and natural products drug discovery. Current science Bangalore 86(6): 789-799.
- Newman DJ, Cragg GM, Snader KM (2003) Natural products as sources of new drugs over the period 1981-2002. J Nat Prod 66(7): 1022-1037.
- Li J, Vederas J (2009) Drug discovery and natural products: end of era or an endless frontier? Science 325(5937): 161-165.
- Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87(1): 43-48.
- Gross H,&Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26(11): 1408-1446.
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringaepv. tomato DC3000. Proc Natl Acad Sci USA 100(18): 10181-10186.
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, et al. (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23(7): 873-878.
- Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, et al. (2005) Comparison of the complete genome sequences of Pseudomonas syringaepv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci USA 102(31): 11064-11069.
- de Bruijn I, de Kock MJ, Yang M, de Waard P, van Beek TA, et al. (2007) Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol 63(2): 417-428.
- Begum AS, Basha AS, Govardhanam R, Kumar MVN, Singh Y, et al. (2014) Isolation and characterization of Antimicrobial cyclic dipeptides from Pseudomonas fluorescens and their efficacy on Sorghum grain mould fungi. Chem Biodivers 11(1): 92-100.
- Khan R, Basha A, Goverdhanam R, Rao PC, Tanemura Y, et al. (2015) Attenuation of TNF-a secretion by l-proline-based cyclic dipeptides produced by culture broth of Pseudomonas aeruginosa. Bioorg Med Chem Lett 25(24): 5756-5761.
- Lindeberg M, Myers CR, Collmer A, Schneider DJ (2008) Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Molecular Plant- Microbe Interactions 21(6): 685-700.
- Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24(5): 1073-1109.
- Chung BS, Aslam Z, Kim SW, Kim GG, Kang HS, et al. (2008) A bacterial endophyte, Pseudomonas brassicacearum YC5480, isolated from the root of Artemisia sp. producing antifungal and phytotoxic compounds. The Plant Pathology Journal 24(4): 461-468.
- Oinuma KI, Hashimoto Y, Konishi K, Goda M, Noguchi T, et al. (2003) Novel Aldoxime Dehydratase Involved in Carbon-Nitrogen Triple Bond Synthesis of Pseudomonas chlororaphis B23 sequencing, gene expression, purification, and characterization. Journal of Biological Chemistry 278: 29600-29608.
- Kamal A, Shaik AB, Kumar CG, Mongolla P, Rani PU, et al. (2012) Metabolic profiling and biological activities of bioactive compounds produced by Pseudomonas sp. strain ICTB-745 isolated from Ladakh, India. J Microbiol Biotechnol 22(1): 69-79.
- Rupesh KR, Priya AM, Prashant K, Jayachandran S (2012) Inhibitory effects of bioactive leads isolated from Pseudomonas aeroginosa PS3 and pseudomonas fluorescence PS 7 on MAP kinases and downregulation of proinflammatory cytokines (TNF a and IL-1 ß) and mediators (NO, iNOS and COX). Toxicol In vitro 26: 571-578.






























