Super-Resolution as a Tool to Understand Nuclear Organization
Rohini R Nair*
Chromatin Dynamic Unit, University of San Raffaele, Italy
Submission: April 23, 2017; Published: April 28, 2017
*Corresponding author: Rohini R Nair, Chromatin Dynamic Unit, University of San Raffaele, Milan, Italy, Email: rohini.nair07@gmail.com
How to cite this article: Rohini R N. Super-Resolution as a Tool to Understand Nuclear Organization. Curr Trends Biomedical Eng & Biosci. 2017; 3(4): 555620. DOI: 10.19080/CTBEB.2017.03.555620
Introduction
Light microscopy has allowed the visualization of cellular and subcellular structures in living and fixed samples. This non- invasive method allows the use fluorescent probes facilitating high contrast and high molecular specificity. However the diffraction limit of the light microscope prohibits in resolving the structure smaller than ~200-300nm.
Chromatin is made up of DNA wrapped around the nucleosomes which are formed by core histone protein H2A, H2B, H3 and H4. In vitro studies, have apparently established the hierarchical model of chromatin compaction, where 10¬nm beads-on-a string chromatin fibres fold into 'higher-order' structures that are ~30nm in diameter which further compact to form 100-200nm chromonema structure [1-3]. Understanding the structure of chromatin is essential since it not only functions in assisting the six feet of DNA to condense and fit inside the nucleus but also assists in gene expression and prevents the DNA from being damaged [4]. However with the spatial resolution of ~200-300nm of a light microscope it is not feasible to study chromatin structure.
The introduction of super-resolution microscope has been able to overcome the diffraction limit extending the spatial resolution of light microscope to length scale as small as 10¬20nm. Super-resolution microscopy methods can broadly be categorized in two types. Structured Illumination Microscopy (SIM) and Stimulated Emission Depletion (STED) takes advantage of patterned illumination light to improve spatial resolution of light microscope. Stochastic Optical Reconstruction Microscopy (STORM) and (Fluorescence) Photo activation Localization Microscopy- PALM and fPALM take advantage of single molecule detection and localization to break the diffraction limit hence both are also referred as Single Molecule Localization Microscopy (SMLM). There are relative benefits and limits of super-resolution microscopy mentioned in Table 1 adapted from Jennifer et al. [5].
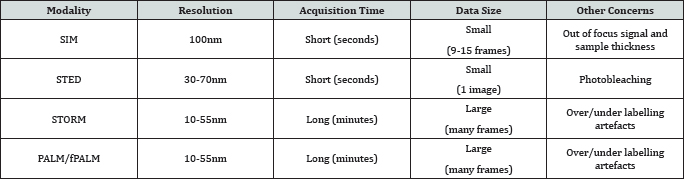
Super-resolution has been extremely useful technique to study chromatin structure. SIM was the first super-resolution method to visualize nuclear structures [6]. Using this approach visualization of chromatin of nuclear pore complex and of nuclear lamina was achieved. It made the precise measurement of chromatin components at higher resolution. SIM was used to further study nuclear compartmentalization and nuclear function [7]. However the SMLM enable for the first time to image histone at high resolution within the intact nuclei [8]. SMLM has also been used in visualizing chromatin plasticity by labelling histones at the nanoscale level as a function of cell state such as pluripotent or differentiated state [9]. Apart from histones SLMN was also used to image DNA [10].
Super-resolution imaging techniques have made it possible to evaluate the organization and dynamics of chromatin at single cell level both in inter phase [8,11] and dividing cell nuclei. It also facilitates to understand the organization of single or groups of nucleosomes and the overall nucleosome occupancy level of DNA [12]. Using quantitative approach the authors identified a new model of chromatin assembly. The results stated that nucleosomes arranged in heterogeneous groups, which the authors named “nucleosome clutches” which were separated by nucleosomedepletedregions.
Hence, super-resolution imaging of histones, DNA RNA polymerase and other nuclear structures in living cells is certainly one of the most interesting fields in the next coming years. I will enable to understand the function of genes at single cell level. However more quantitative methods have to be developed to builds predictive model about gene function. Hence, using super-resolution microscopy it not far to view the activity of gene at single cell level.
References
- Finch JT, Klug A (1976) Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA 73(6): 1897-1901.
- Song F, Chen P, Sun D, Wang M, Dong L, et al. (2014) Cryo-EM study of the chromatin fiber reveals a double helixtwisted by tetranucleosomal units. Science 344: 376-380.
- Widom J (1992) A relationship between the helical twist of DNA and the orderedpositioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci USA 89: 1095-1099.
- Lavelle C (2014) Pack, unpack, bend, twist, pull, push: the physicalside of gene expression. Curr Opin Genet Dev 25: 74-84.
- Jennifer A, Thorley, Jeremy Pike, Joshua Z, Rappoport (2014) Superresolution Microscopy: A Comparisonof Commercially Available Options. Fluorescence Microscopy. Elsevier 2014: 199-212.
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, et al. (2008) Subdiffractionmulticolor imaging of the nuclear periphery with 3Dstructured illumination microscopy. Science 320(5881): 13321336.
- Markaki Y, Gunkel M, Schermelleh L, Beichmanis S, Neumann J, (2010) Functional nuclear organization of transcription and DNA replication: atopographical marriage between chromatin domains and the inter chromatin compartment. Cold Spring Harb Symp Quant Biol 75: 475492.
- Bohn M, Diesinger P, Kaufmann R, Weiland Y, Muller P (2010) Localization microscopy reveals expression-dependent parameters of chromatin nanostructure. Biophys J 99: 1358-1367.
- Ricci MA, Manzo C, Garci'a-Parajo MF, Lakadamyali M, Cosma MP (2015) Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160: 1145-1158.
- Zessin PJ, Finan K, Heilemann M (2012) Super-resolution fluorescenceimaging of chromosomal DNA. J Struct Biol 177: 344-348.
- Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, et al. (2010) Live-cell super-resolution imaging withtrimethoprim conjugates. Nat Methods 7: 717-719.
- Matsuda A, Shao L, Boulanger J, Kervrann C, Carlton PM, et al. (2010) Condensed mitotic chromosome structure atnanometer resolution using PALM and EGFP-histones. PLoS One 5(9): e12768.






























