Synthetic DNA Crown Cells Kill Yeast: Relationship with Assembly Formation of Yeast
Shoshi Inooka*
Japan Association of Science Specialists, Japan
Submission: August 17, 2021; Published: August 27, 2021
*Corresponding Author: Shoshi Inooka, Japan Association of Science, Japan
How to cite this article: Shoshi I. Synthetic DNA Crown Cells Kill Yeast: Relationship with Assembly Formation of Yeast. Civil Eng Res J. 2021; 12(2): 555831. DOI 10.19080/CERJ.2021.12.555831
Abstract
DNA crown cells are artificial cells in which the outside of the membrane is coated with DNA. Synthetic DNA crown cells are readily prepared from mixtures of sphingosine (Sph), DNA, and adenosine. and can multiply within suspensions of egg white, yielding cells that also are referred to as DNA crown cells. To date, many kinds of both types of DNA crown cells have been prepared using various DNAs. However, it remains unclear whether DNA crown cells are biologically relevant. During my studies of DNA crown cells, I showed that antibiotics are produced in co-cultures of DNA crown cells with brewer’s yeast. This observation suggested that DNA crown cells may control the growth of yeast. In subsequent work to clarify that issue, I showed that assemblies are formed in mixtures of synthetic DNA (Escherichia. coli) crown cells with Bacillus subtilis, resulting in inhibition of the growth of B. subtilis. In the present work, synthetic DNA crown cells were generated using three kinds of DNA (E. coli, Streptomyces sp., and akoya pearl oysters) and tested for effects on brewer’s yeast. The results showed that assemblies of yeast were formed in the co-cultures of synthetic DNA crown cells and yeast, such that the yeast were unable to grow, meaning that synthetic DNA crown cells kill yeast. Finally, I discuss the mechanism whereby the binding of synthetic DNA crown cells to yeast may result in the formation of yeast assemblies and killing of the yeast cells.
Keywords: Synthesized DNA crown cells; Sphingosine-DNA; Killing of yeast; Assembly of yeast
Introduction
There has been significant progress in the generation of artificial cells. Recently, approaches for generating fully operational (self-replicating) artificial cells have been reported [1,2]. Such artificial cells consist of membranes covered with DNA (named DNA crown cells) [3] and are generated when these DNA-coated membranes are incubated with egg white. DNA crown cells are readily synthesized as follows: DNA is added to sphingosine (Sph). After the heating of Sph-DNA mixtures, A-M compounds (consisting of mixtures of adenosine and monolaurin) are added to the mixtures. DNA crown cells prepared by such methods are called synthetic DNA crown cells. On the other hand, DNA crown cells that are prepared when synthetic DNA crown cells are incubated with egg white are called DNA crown cells. It may be important to distinguish between DNA crown cells before multiplication (synthetic DNA crown cells) and after multiplication (DNA crown cells) because there may be differences in function between the two types of cells. Several kinds of DNA crown cells have been prepared, both before and after multiplication [4-7]. On the other hand, there is no evidence whether such DNA crown cells have some biological functions. In previous experiments [8], it was reported that antibiotics are produced in co-cultures of DNA crown cells (prepared using DNA from Streptomyces griseus) with brewer’s yeast, and that the growth of yeast that are co-cultured with DNA crown cells is suppressed, although DNA crown cells may continue to grow. Moreover, previous experiments [9] demonstrated that synthetic DNA crown cells were formed when mixed with Bacillus subtilis, such that these crown cells suppressed the growth of B. subtilis. The present experiments were carried out to clarify whether the formation of cell assemblies and the suppression of B. subtilis growth, which was observed in the combination of synthetic DNA (E. coli) crown cells with B. subtilis, also was observed in combinations of yeast with each of three kinds of synthetic DNA (E. coli, Streptomyces sp., and akoya pearl oyster) crown cells. In the start of the present study, three kinds of DNA crown cells were synthesized with Sph-DNA-A-M mixtures as described previously [10]. Then, the resulting synthetic DNA crown cells were mixed with yeast. Following mixing, flat assemblies, which consisted of many yeast cells, were formed. Moreover, after the mixture of synthetic DNA crown cells and yeast was incubated for 3 or 24 hours, respectively, at 37 °C, yeast viability was assessed. Notably, no viable yeast was observed at either 3 or 24 hours, respectively, suggesting that the yeast died. The formation of the assemblies and killing of the yeast may be based on the binding of synthetic DNA crown cells to yeast, given that such binding was observed when these mixture were evaluated by microscopy. Thus, the present experiments demonstrated that large flat assemblies consisting of many yeast cells were formed when synthetic DNA crown cells were mixed with yeast, leading to the killing of this yeast within 3 hours. The formation of the large flat assemblies with yeast and killing of yeast were observed in mixtures of each of the three kinds of synthetic DNA crown cells with yeast. Therefore, these abilities may be general functions associated with synthetic DNA crown cells. On the other hand, it is unclear whether synthetic DNA crown cells live or multiply in the mixtures containing yeast. In a previous report [8] on the mechanism of antibiotic production in co-cultures of DNA crown cells (not synthetic DNA crown cells) with brewer’s yeast, it was proposed that DNA crown cells may multiply. Further potential functions of synthetic DNA crown cells are expected to be identified in experiments testing whether synthetic DNA crown cells multiply in mixtures containing yeast and whether such cells kill yeast.
Materials and Methods
Materials
The following materials were used:
Sph was obtained from (Sigma, USA, and Tokyo Kasei, Japan). Three kinds of DNA were used, as follows. E. coli DNA (B1 strain) was purchased commercially from Sigma, USA. DNA from Streptomyces sp. and from akoya pearl oysters was extracted [5,8]. Adenosine (Sigma, USA, and Wako, Japan) and monolaurin (Tokyo Kasei, Japan) were obtained commercially. A-M compound was synthesized from mixtures of adenosine and monolaurin as described previously [10]. Yeast was obtained as dry ale yeast (Safale S-04) from Formentis, Bergy. For use in the experiments, dry yeast was suspended in distilled water to a density of approximately 108 cells per mL, as determined by counting with a hemocytometer. The culture media for yeast were as follows. Sanisupetsuk raw medium (Azuwan, Japan) was used to test yeast viability. YM medium consisted of Difco YM Broth (Becton Dickinson, MD, USA) supplemented with agar. Nutrient Agar (NA) medium (Oxoid, UK) was used to separate yeast.
Methods
a. Preparation of DNA crown cells
Here, three kinds of DNA crown cells were synthesized using virtually identical methods, albeit with slightly different concentrations of DNA. Synthetic DNA crown cells were prepared as described previously [9]. Briefly, l80 μg of Sph (10 mM) and 90 μL of DNA (approximately 1.7 μg/μL) were mixed, and the mixture then was heated and cooled. These procedures were carried out twice. Next, A-M compound (100 μL) was added, and the mixture was incubated at 37 °C for 15 minutes. Then, 30 μL of monolaurin were added and the mixture was incubated at 37 °C for 5 minutes. The resulting mixture was used as a suspension of synthetic DNA crown cells.
b. The mixture of yeast with synthetic DNA crown cells
An aliquot (50 μL) of yeast suspension was added to 50 μL of synthetic DNA crown cells and the combination was mixed. These samples were prepared in duplicate, with one each incubated for 3 hours and 24 hours. At study start, a pre-study aliquot of each mixture was removed and used as a sample without incubation. The remainder of the mixtures then were incubated for 3 hours or 24 hours at 37 °C.
c. Microscopic observation
A drop of the synthetic DNA crown cells that had been cultured with yeast for 0.5, 1.5. 3.0 and 24 hours, respectively, were placed on a slide glass and covered with a cover glass. This slide then was observed by light microscopy.
d. Viable yeast in the mixtures of synthetic DNA crown cells The following samples were tested.
a) The sample after the incubation for 3 hours at 37 °C
b) The sample after the incubation for 24 hours at 37 °C
c) The sample after the incubation for 3 and 24 hours at 37 °C of yeast without crown cells.
At the indicated time points, 50 μL of each sample was spread on agar plates and incubated for 18 hours at 37 °C. Colonies that grew on these plates were counted and used to calculate viability.
e. Attempt to isolate yeast from the mixtures with synthetic DNA (Streptomyces) crown cells
An aliquot of 50 μL of synthetic DNA crown cells was added to 50 μL of yeast and the mixtures were incubated for 24 hours at 37 °C. A portion of the mixture then was transported to Techno Suruga Lab, where the isolation of yeast from the sample were performed under the following culture conditions (a and b):
a) For 7~14 days at 27 °C in YM medium;
b) For 7 and 14 days at 27 °C and 37 °C, respectively, in NA medium.
Results and Discussion
Microscopic appearance of synthetic DNA crown cells and yeast
Microscopic appearance of synthetic DNA crown cells
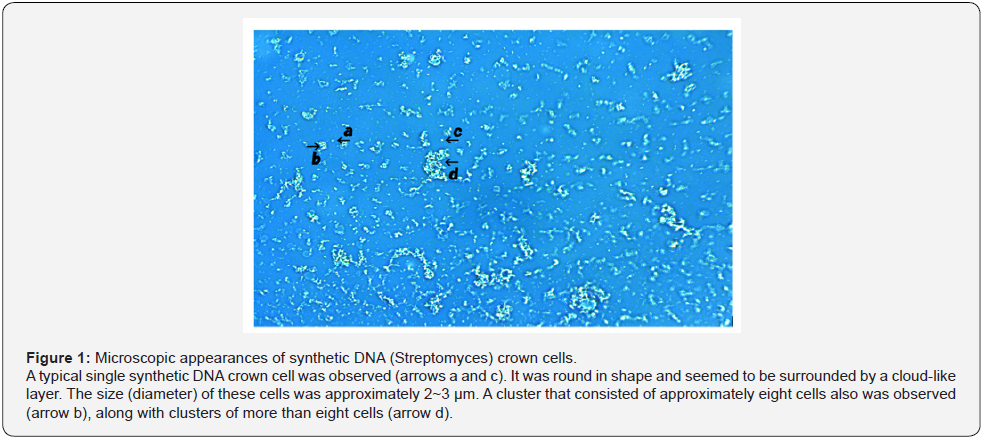
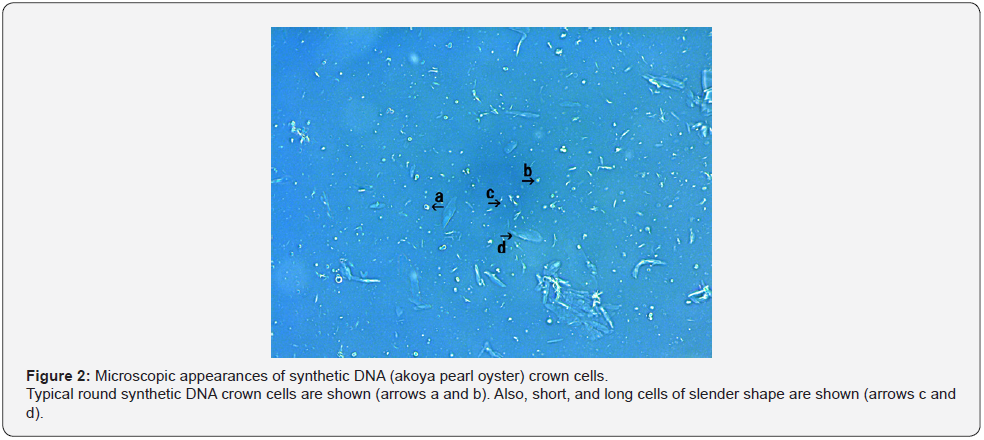
Three kinds of DNA crown cells were prepared using DNA (obtained commercially) from E. coli and DNA extracted inhouse from Streptomyces sp. and from akoya pearl oyster. Each kind of DNA crown cells appeared similar in shape and size and was similar to the microscopic appearance described previously for synthetic DNA (E. coli) crown cells [11]. Therefore, synthetic DNA (Streptomyces) crown cells and DNA (akoya pearl oyster) crown cells were similar to the previously described phenotype. Synthetic DNA (Streptomyces) crown cells are shown in Figure 1. A typical single DNA crown cell is shown in Figure 1 (arrows a and c). This cell was round in shape and seemed to be surrounded by a cloud-like layer. The size of a typical cell was approximately 2~3 μm in diameter. Clusters consisting of multiple DNA crown cells (approximately 8 cells) also were observed (Figure 1, arrow b), as well as super-clusters consisting of several clusters of DNA crown cells (Figure 1, arrow d). Most of objects shown in Figure 1 (not indicated by arrows) constituted single cells or clusters of single cells. Synthetic DNA (akoya oyster pearl) crown cells ae shown in Figure 2. Typical round cells are indicated by arrows a and b. Also, a cell with a slender-like shape is indicated by arrow c. The surface of the cells indicated by arrows b and c consisted of cloud-like substances. Most of the objects shown in Figure 2 appeared to be DNA crown cell of various sizes with round or slender profiles (Figure 2, arrow d). Here, two kinds of synthetic DNA crown cells were introduced, and synthetic DNA (E. coli) crown cells were as previously described [11]. Each cell appears to possess a characteristic shape or size. On the other hand, their characteristics in shape or size may be determined by the conditions use to prepare such cells, such as the purity of DNA or Sph, heating of Sph-DNA fibers, and so on. Accordingly, it may be difficult to prepare cells that are consistent in size or shape between experiments. However, the general resemblance among different kinds of crown cells suggested a common structure in which the outside of cells consists of Sph-DNA, regardless of the condition used for preparation or the type of DNA.
Microscopic observation of yeast
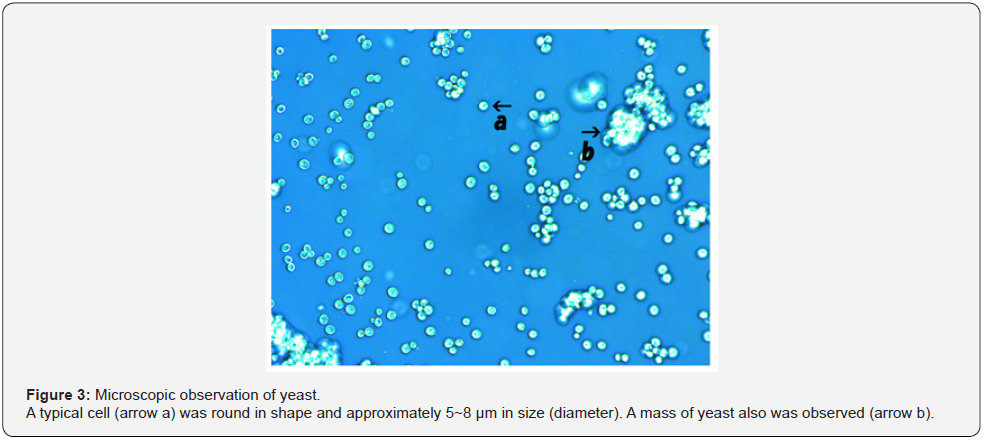
Yeast that was used in the present experiments are shown in Figure 3. A typical cell (Figure 3, arrow a) was round in shape and approximately 5~8 μm in size (estimated by scale bar, as shown in Figure 11a). In this sample, a clump of yeast was observed (Figure 3. arrow b).
The formation of assemblies in mixtures of synthetic DNA crown cells with yeast
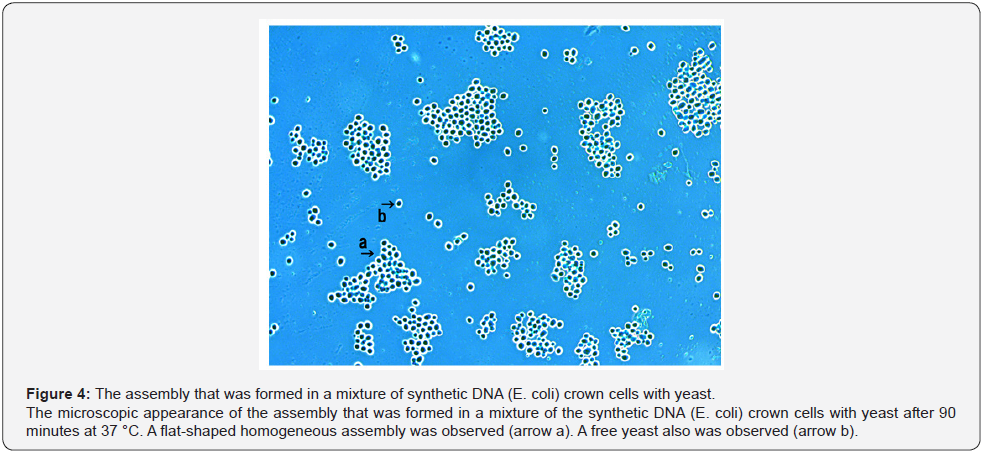
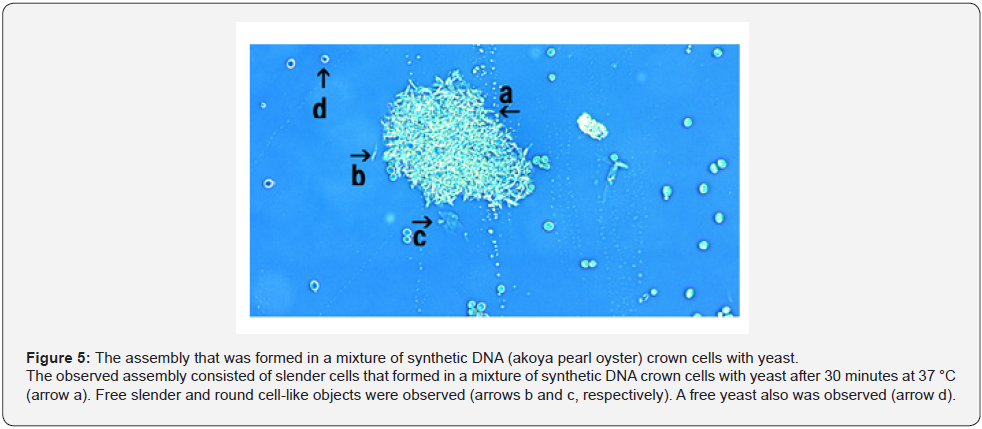
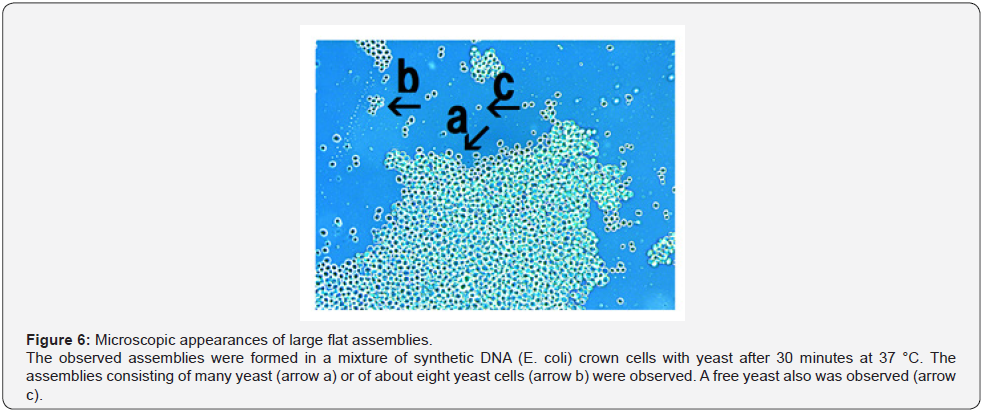
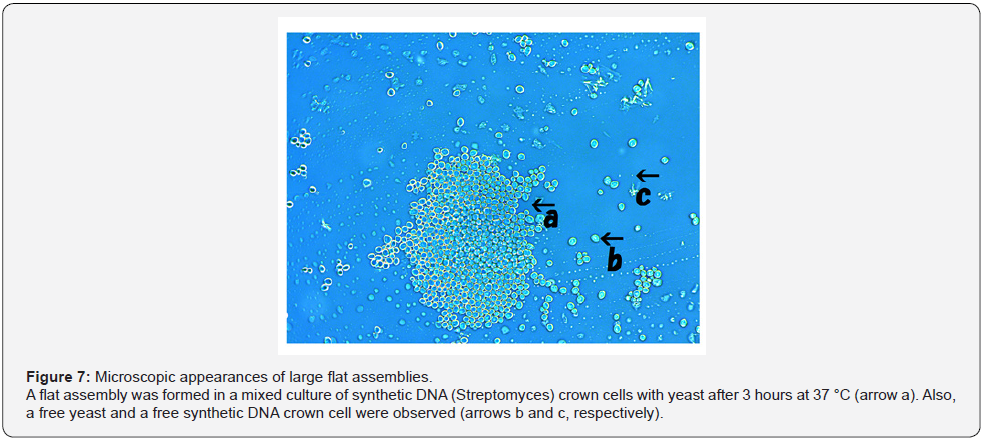
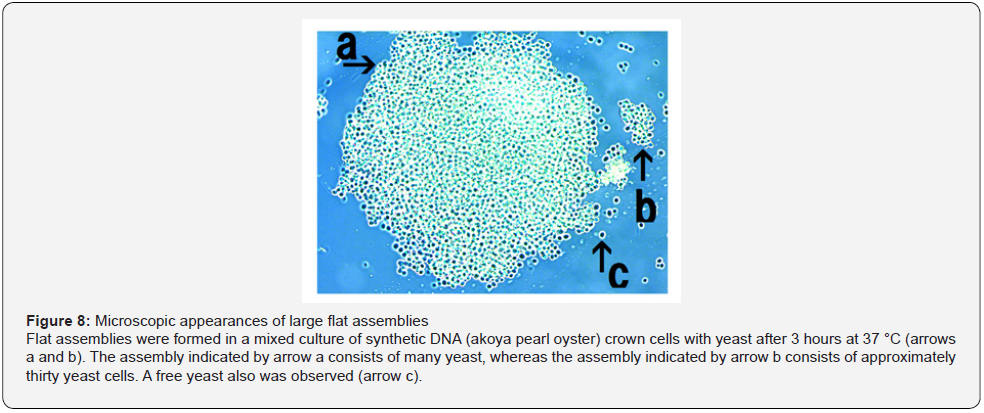
When yeast was added to the DNA crown cells, two basic kinds of the assemblies (flat and fibrous) were observed. Figure 4 shows the appearance of the assemblies formed by mixtures of the synthetic DNA (E. coli) with yeast after incubation for 90 minutes at 37 °C. Assemblies of various sizes and shapes were observed (Figure 4, arrow a). Here, such assemblies are designated flat assemblies. The appearances of the assembly that was observed after the mixture of synthetic DNA (akoya pearl oyster) crown cells with yeast was incubated for 30 minutes at 37 °C is shown in Figure 5 (arrow a). A slender assembly of free cells was observed (as shown in Figure 5, arrow b). This assembly may consist of a mass of cell-like slender domains. Here, such assemblies are called fibrous assemblies. A free yeast was observed (Figure 5, arrow d) and free round cells also were observed (Figure 5, arrow c). For the culture of yeast, the flat assembly was observed more frequently than the fibrous assembly. In previous reports [11,12], it was noted that assemblies of various shapes and sizes formed when synthetic DNA (E. coli and human placenta) crown cells were added to B. subtilis. Based on their microscopic appearance, flat assemblies appear to be formed via very simple and loose binding. These results suggested that the makeup of the assemblies differed depending on the combination of synthetic DNA crown cells and bacteria used. On the other hand, large flat assemblies of various sizes were observed consistently for all combinations of synthetic DNA crown cells with yeast (Figures 4, 5 & 6). The assemblies that were formed from mixtures of synthetic DNA (E. coli) crown cells with yeast for 30 minutes at 37 °C is shown in Figure 6. Assemblies consisting of many yeast (Figure 6 arrow a) and of about eight yeast (Figure 6, arrow b) were observed. A free yeast also was observed (Figure 6 arrow c). The flat assembly that was formed in mixed cultures of synthetic DNA (Streptomyces) crown cells with yeast for 3 hours at 37 °C is shown in Figure 7, arrow a, Also, a free yeast and a free synthetic DNA crown cell were observed (Figure 7, arrows b and c, respectively). The flat assembly that was formed in mixed cultures of synthetic DNA (akoya oyster pearl) crown cells with yeast for 3 hours at 37 °C is shown in Figure 8 (arrows a and b). The assembly indicated by arrow a consisted of many yeast cells, which were too numerous to count, whereas the assembly indicated by arrow b consisted of approximately thirty yeast. A free yeast also was observed (Figure 8, arrow c). Because the large flat assemblies of yeast were obtained with each of the three tested kinds of synthetic DNA crown cells, I infer that synthetic DNA crown cells possess the potential to form large flat assemblies. The formation of the large assembly appeared to be influenced by culture (temperature), given that the formation of assemblies was observed before incubation. Specifically, the formation of the assembly occurred out as soon as the synthetic DNA crown cells were mixed with yeast. The sizes of assemblies were not assessed in the present set of experiments, given that the exact size of assemblies may be not important. By the way, a typical single yeast is 5~8 μm as indicated in Figure 11a.
Survival of yeast following mixed cultures with synthetic DNA crown
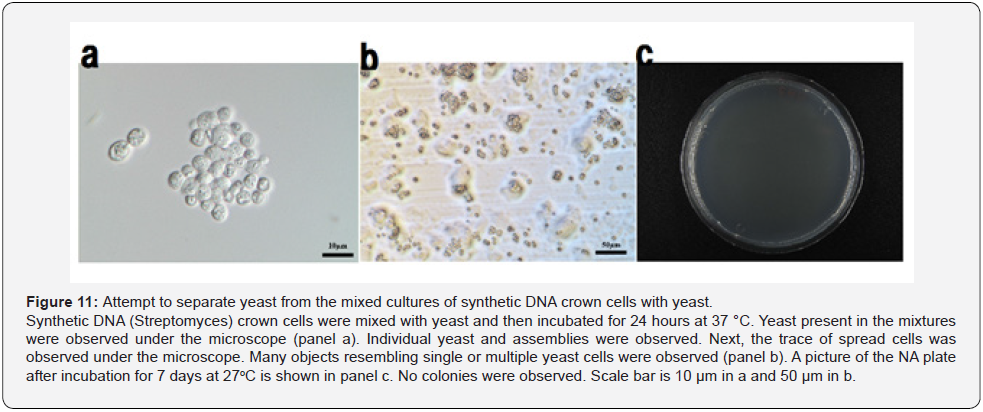
After mixtures of synthetic DNA (E. coli) crown cells with yeast were cultured at 37 °C for 3 or 24 hours, aliquots were plated and incubated at room temperature (approximately 20 °C) for 3~5 days. No viable yeast was observed from either culture (Figure 9). The same experiments were carried out using synthetic DNA (Streptomyces and akoya pearl oyster) crown cells. Again, no viable yeast was observed (Figure 10). The findings showed that yeast cultured in the presence of synthetic DNA crown cells lost viability. These results indicated that all of the yeast in large flat assemblies died; presumably, synthetic DNA crown cells killed all of the yeast incorporated into the assembly within 3 hours. To confirm whether yeast in the mixtures with synthetic DNA crown cells grow, further experiments were carried out at another site. Specifically, synthetic DNA (Streptomyces) crown cells were mixed with yeast and then incubated for 24 hours at 37 °C. The sample was transported to Techno Suruga Laboratory Co., Ltd. (Shizuoka, Japan). Here, yeast in the sample were observed by microscopy (Figure 11a). Then, the number of yeast in the sample was counted via hemocytometer, revealing the presence of yeast at a density of approximately 1.3 x 108/ mL. Next, the mixture of synthetic DNA (Streptomyces) crown cells and yeast was transferred to either of two kinds of media. One portion was spread on YM medium, and these plates were and incubated for 7 ~14 days at 27 °C. A second portion was spread on duplicate NA medium plates, which were incubated for 7~14 days at 27 °C or 37 °C. Before incubation, the traces of spread cells were observed under the microscope. As shown in Figure 11b, many objects that may have represented single or multiple yeast cells were observed. Figure 11c shows an image of an NA plate following incubation for 7 days at 27 °C. Notably, no colonies were observed. In these all cultures, no yeast colonies were observed, suggesting that all of plated yeast were dead.
Binding to yeast of synthetic DNA crown cells
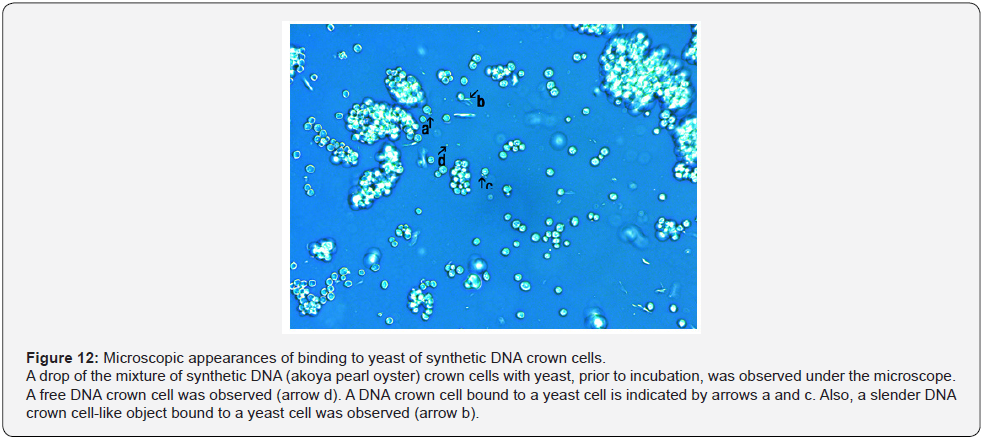
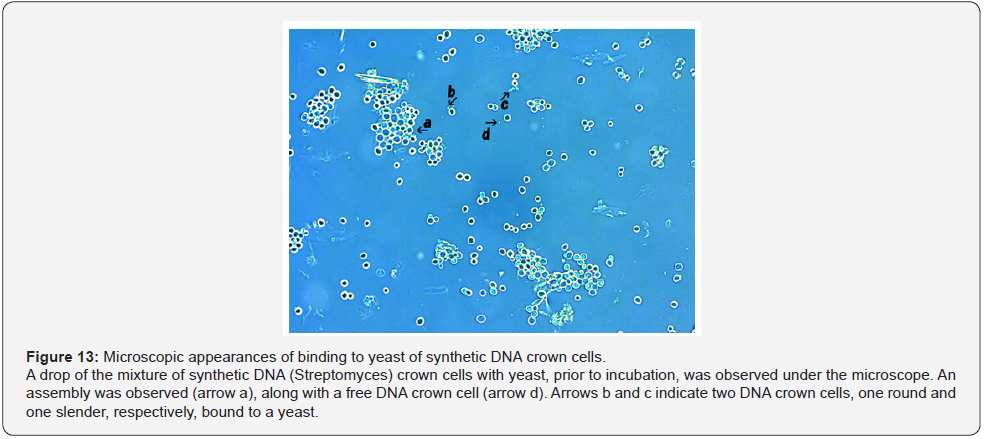
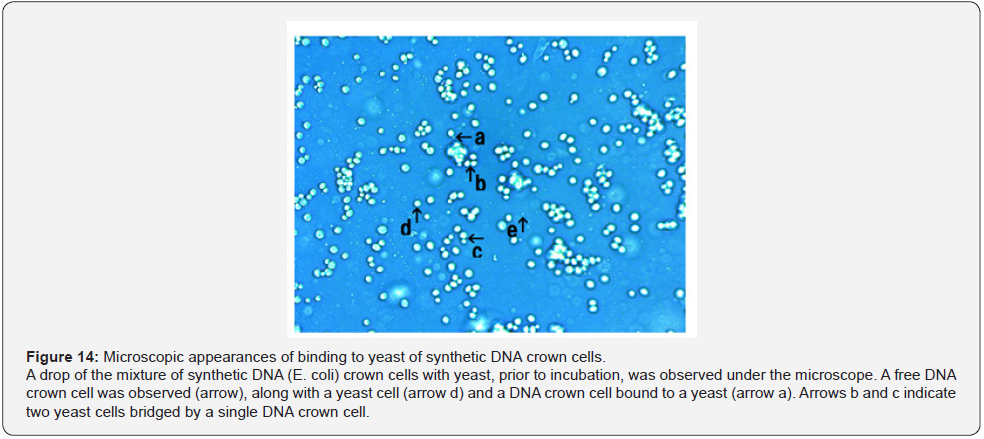
It may be difficult to clarify the mechanisms of the assembly formation, especially that of the large flat assemblies. To obtain evidence, careful microscopic observation was carried out using samples obtained from mixtures of synthetic DNA crown cells with yeast prior to incubation. The microscopic appearances of the mixtures of synthetic DNA (akoya pearl oyster) crown cells with yeast are shown in Figure 12. A round object in the size range expected for a free DNA crown cell was observed (Figure 12, arrow d). This cell was bound to a yeast, as shown in Figure 12, arrows a and c. Also, a slender cell-like object was seen bound to a yeast (Figure 12. arrow b). Figure 13 shows the microscopic appearances of the mixtures of synthetic DNA (Streptomyces) crown cells with yeast. An assembly was observed in Figure 13, arrow a. A round object in the size range expected for a free DNA crown cell was observed (Figure 13, arrow d). Arrows b and c in Figure 13 indicate two DNA crown cell-like objects, one round and one slender, respectively, both bound to a yeast. Figure 14 shows the microscopic appearances of mixtures of synthetic DNA (E. coli) crown cells with yeast. A free DNA crown cell was observed in Figure 14, arrow e, and a free yeast was observed in Figure 14, arrow d. Another two DNA crown cells bound to a yeast were observed (Figure14, arrow a). A DNA crown cell was observed between two yeast cells (Figure 14. arrows b and c), suggesting that yeast are linked via bridges consisting of DNA crown cells. Thus, the present experiments demonstrated that flat assemblies are formed between synthetic DNA crown cells and yeast, and that synthetic DNA crown cells kill yeast within 3 hours of exposure. It previously has been reported [10,12] that assemblies of various shapes and sizes are formed in mixtures of synthetic DNA (E. coli and human placenta) crown cells and B. subtilis. Such assemblies with B. subtilis may be constructed primarily with DNA crown cells. On the other hand, when crown cells were combined with yeast, homogeneous assemblies that are flat in shape are the primary product and may consist primarily of the standard cells (yeast). The facts suggested that there are assemblies that are formed using DNA crown cells as the main material and assemblies that are formed using bacteria (or yeast) as the main material. The formation of the flat assemblies may begin from the binding of synthetic DNA crown cells to yeast. As shown in Figures 12, 13 & 14, it is clear that yeast bind to three kinds of synthetic DNA crown cells. On the other hand, the micrographs in Figure 14 showed what appeared to be two yeast cells bridged by an object resembling a DNA crown cell. Now, although there was not sufficient evidence to clearly show that yeast is bridged by DNA crown cells, I conjecture that synthetic DNA crown cells form a physical link between yeast cells, leading to the formation of large flat assemblies.
A previous report [9] tested whether the growth of B. subtilis is suppressed by synthetic DNA (E. coli) crown cells, showing that only a small number of B. subtilis survived in such mixtures. Regarding the mechanisms of growth suppression, it was suggested that B. subtilis were enclosed within assemblies, thereby suppressing growth of these bacteria. When synthetic DNA crown cells were combined with yeast, no growth of yeast was observed. This result indicated that all yeast incorporated into large assemblies are killed by synthetic DNA crown cells within 3 hours. These facts suggested that killing of yeast may be associated with the assembly formation, and may begin with the binding of synthetic DNA crown cells to yeast. Moreover, the binding may occur between Sph on outside of synthetic DNA crown cells and a component of the yeast cell wall. On the other hand, the fate of synthetic DNA crown cells in mixtures with yeast remains unclear. Initial studies on artificial cells [13] reported that Sph- DNA exhibits cytotoxicity against cultured L-M cells. In short, L-M cells were destroyed when Sph-DNA particles were added to the cells and viable particles (artificial cells) were generated [10,13]. If such a case also applies for yeast, as for L-M cells, it would appear that synthetic DNA crown cells acquire living-ability after destroying yeast, subsequently multiplying in the mixed cultures with yeast. Thus, here, it was demonstrated that synthetic DNA crown cells kill yeast after 3 hours in mixed culture. The present findings may indicate a potential function possessed by synthetic DNA crown cells.
Acknowledgment
I thank Emi Miwa (Techno Suruga Laboratory, Co. Ltd., Shizuoka, Japan) for assistance in cloning yeast and for useful discussions.
References
- Inooka S (2012) Preparation, and cultivation of artificial cells. App Cell Biol 25: 13-18.
- Inooka S (2016) Preparation of Artificial Cells Using Eggs with Sphingosine-DNA. J Chem Eng Process Technol l7: 277.
- Inooka S (2016) Aggregation of sphingosine-DNA and cell construction using components from egg white. Integrative Molecular Medicine 3(6): 1-5.
- Inooka S (2017) Systematic Preparation of Bovine Meat DNA Crown Cells App Cell Biol Japan 30:13-16.
- Inooka S (2018) Systematic Preparation of DNA (Akoya pearl oyster) Crown Cells App Cell Bio Japan 31: 21-34.
- Inooka S (2019) Preparation of DNA (Nannochloropsis sepsis) Crown Cells Using eggs, sphingoshin and DNA, and subsequent cell recovery App Cell Biol Japan 32: 55-64.
- Inooka S (2014) Preparation of Artificial Human Placental Cells App. Cell Biol. 27: 43-49.
- Inooka S (2020) On the mechanism of Antibiotic Production in Co-Cultures of Generated DNA (Streptomyces Griseus) Crown Cells (Artificial Cells) with Yeast (Beer). Novel Research in Science 5(3): 1-5.
- Inooka S (2021) Growth suppression of Bacillus Subtilis in Co-cultures with DNA (Escherichia Coli) Crown Cells, Including Possible Bacterial Transformation within the Assemblies. Bioscience and Biotechnology Journal 2(1): 22-25.
- Inooka S (2017) Biotechnical and Systematic Preparation of Artificial Cells (DNA Crown Cells). The Global Journal of Research in Engineering 17(1-C).
- Inooka S (2021) The assembly formation by co-cultures of DNA (Escherichia coli) crown cells with Bacillus subtilis cells. Journal of Biotechnology & Bioresearch 2(5): 1-5.
- Inooka S (2021) Microscopic observation of assemblies formed in mixtures of DNA (human placenta) crown cells with Bacillus subtilis. An archive of organic and in-organic chemical sciences 5(2): 658-664.
- Inooka S (2000) Cyto- organisms (cell-originated cultivable particles) with Sphingoshin-DNA. Comm Appl Cell Biol 17: 11-34.






























